#Install these packages first
# install.packages(c("sf","elevatr","terra","stars","tidycensus"))
# install.packages('devtools')
# devtools::install_github("ropensci/USAboundaries")
# install.packages("USAboundariesData", repos = "https://ropensci.r-universe.dev", type = "source")
library(tidyverse)
library(sf) # tools for working with spatial vector data (GIS functionality, mapping)
library(elevatr) # access to raster elevation maps
library(terra)
library(stars)
library(tidycensus) # spatial data for the US with census information
library(USAboundaries) # access to boundaries for US states, counties, zip codes, and congressional districts 4 Advanced Spatial Visualization - Part 1
Settling In
Data Storytelling Moment
Learning goals
After this lesson, you should be able to:
- Understand the basics of a CRS (coordinate reference system)
- Understand and recognize different spatial file types and data types in R
- Implement some of the basic plotting with the
sfpackage - Understand foundational ideas in working with spatial data (aggregating spatial point data to a spatial region, joining spatial data sets)
Additional resources
- Spatial Data Science https://r-spatial.org/book/
- Spatial Data Science with R and
terra: https://rspatial.org/ - Leaflet in R https://rstudio.github.io/leaflet/
- CRAN task view on spatial analysis: https://cran.r-project.org/web/views/Spatial.html
Setup
Create the following directory structure for this activity:
04_adv_mapscode04-adv-maps-1-notes.qmd
data- We will be saving data here during this activity.
figures- We will be saving created maps here during this activity.
First load required packages.
Spatial Data in R
See Spatial data notes for basics of CRS and spatial data types.
Download Shapefiles
- Navigate to the following URLs to download the spatial data files we’ll be using in this activity. Put these files in the
datafolder of your04_adv_mapsfolder.
- MN cities: https://gisdata.mn.gov/dataset/loc-pop-centers
- File type: shapefile (
.shp) - File name:
shp_loc_pop_centers.zip(Unzip this after downloading.)
- File type: shapefile (
- MN water: https://gisdata.mn.gov/dataset/us-mn-state-metc-water-lakes-rivers
- File type: shapefile (
.shp) - File name:
shp_water_lakes_rivers.zip(Unzip this after downloading.)
- File type: shapefile (
Read in Files
- Read in the MN cities and MN water shapefiles by entering the correct relative paths in
st_read(). Tab completion will be very helpful here: type part of a directory or file name and hit tab to autocomplete or bring up a dropdown of options.
# The sf package comes with a North Carolina shapefile:
nc <- st_read(system.file("shape/nc.shp", package = "sf"))
# Read in shapefiles just downloaded
mn_cities <- st_read("Add correct relative path to city_and_township_population_centers.shp")
mn_water <- st_read("Add correct relative path to LakesAndRivers.shp")The sf package reads in spatial data in data.frame-like format. Using the class() function we can check the class (type) of object that we just read in. Note the presence of the “sf” and “data.frame” classes:
class(nc)
class(mn_cities)
class(mn_water)When we read in spatial objects, it is useful to check what CRS underlies the data. We can do that with st_crs() from the sf package:
st_crs(nc)We can treat sf objects similarly to ordinary datasets when using ggplot2 to make spatial visualizations:
ggplot(nc) +
geom_sf() +
theme_classic() +
labs(title = "NAD27")
Change CRS
- Let’s explore how changing the CRS changes the map. The
st_transform()function insfre-expresses a spatial object using a user-supplied CRS. Thecrsargument takes a string descriptor of the CRS. We can find these descriptors via https://epsg.io. In the example below, I searched for “South Carolina”.
nc_transformed <- nc %>% st_transform(crs = "EPSG:32133")
st_crs(nc_transformed)
ggplot(nc_transformed) +
geom_sf() +
theme_classic()The goal is to use https://epsg.io to find two CRSs that result in a North Carolina map that is noticeably different from the original in the NAD27 CRS.
Take a look at the function below that re-maps a spatial object using a new CRS.
- Read through the function to get a sense for how this code works.
spatial_objandnew_crsare called arguments (function inputs).- Add one more argument called
titleto this function. Use this input to set the plot title.
- Add one more argument called
- Use your function to make two new maps using your chosen CRSs.
transform_and_plot <- function(spatial_obj, new_crs) {
spatial_obj %>%
st_transform(crs = new_crs) %>%
ggplot() +
geom_sf() +
theme_classic()
}
# Example usage of this function (using a South Carolina CRS)
transform_and_plot(nc, new_crs = "EPSG:32133")Verify your understanding: If you had point location data that was not in the NAD27 CRS, what would you expect about the accuracy of how they would be overlaid on the original North Carolina map?
MN map with multiple layers
Goal: create a map of MN with different layers of information (city point locations, county polygon boundaries, rivers as lines and polygons, and a raster elevation map).
Get county boundaries
- We’ve already read in city location and water information from external shapefiles. We can access county boundaries with the
us_counties()function in theUSAboundariespackage.
# Load country boundaries data as sf object
mn_counties <- USAboundaries::us_counties(resolution = "high", states = "Minnesota")
# Take care of duplicate column names (there are two identical "state_name" columns)
names_counties <- names(mn_counties)
names(mn_counties)[names_counties == "state_name"] <- c("state_name1", "state_name2")
Unifying CRSs across different spatial datasets
- We first need to ensure that the CRS is the same for all spatial datasets.
- Check the CRS for the
mn_cities,mn_water, andmn_countiesdatasets. - If the datasets don’t all have the same CRS, use
st_transform()to update the datasets to have the same CRS asmn_cities. You can usecrs = st_crs(mn_cities)withinst_transform().
Counties + cities
- Create a map where city locations are overlaid on a map of county boundaries.
- You will need to call
geom_sf()twice. - Make the map background white.
- Install the
ggthemespackage, and add the following layer to use a clean map theme:+ ggthemes::theme_map()
Customize Colors
- We can use traditional
ggplot2aesthetics (e.g.,fill,color) to display location specific attributes. Below we only plot large cities, and we color and size cities according to their population.
ggplot() +
geom_sf(data = mn_counties, fill = "white") +
geom_sf(data = mn_cities %>% filter(Population >= 10000), mapping = aes(color = Population, size = Population)) + # cities layer
scale_color_viridis_c() + # continuous (gradient) color scale
labs(title = "Minnesota Cities with Population >= 10,000") +
ggthemes::theme_map() +
theme(legend.position = "bottom") # move legendLook up the scale_color_viridis_c() documentation via the ggplot2 reference.
- Read the function description at the top. What is the advantage of using this function for making color palettes?
- Look through the examples section. What is the difference between the
_d(),_c(), and_b()variants of this function?
Adding elevation raster data
Where are large cities located? Is there some relationship to local geography/terrain?
- To investigate these questions, we can obtain elevation data to include on the map using the
elevatrpackage. We encounter two new functions here—we can look up their documentation to make sense of the code by entering the following in the Console:
?elevatr::get_elev_raster?terra::as.data.frame
elevation <- elevatr::get_elev_raster(mn_counties, z = 5, clip = "bbox")
raster::crs(elevation) <- sf::st_crs(mn_counties)
# Convert to data frame for plotting
elev_df <- elevation %>% terra::as.data.frame(xy = TRUE)
colnames(elev_df) <- c("x", "y", "elevation")Build on our existing map by adding a raster layer for elevation as the background.
- Look up the documentation for
geom_raster()to plot the elevation data fromelev_df. This will be the first layer of the plot. - Look at the documentation for
scale_fill_gradient()to add the following elevation color scale:"darkgreen"represents the lowest elevations, and"white"represents the highest elevations. - Add in the layers from the map above to show the largest cities and the county outlines. To remove a background color, use
fill = NA.
Zoom in to Twin Cities and add water
- The bulk of the interesting information in this map is in the Twin Cities area. Let’s zoom in to this area.
- We can use the
st_bbox()function to get the bounding box for a spatial object—we do this after filtering to the 7 counties in the Twin Cities. - We then use
st_crop()to trim a spatial object to a given bounding box.
seven_countyarea <- mn_counties %>%
filter(name %in% c("Anoka", "Hennepin", "Ramsey", "Dakota", "Carver", "Washington", "Scott")) %>%
st_bbox()
seven_countyarea
elevation <- elevatr::get_elev_raster(mn_counties %>% st_crop(seven_countyarea), z = 9, clip = "bbox")
raster::crs(elevation) <- sf::st_crs(mn_counties)
# Convert to data frame for plotting
elev_df <- elevation %>% terra::as.data.frame(xy = TRUE)
colnames(elev_df) <- c("x", "y", "elevation")In the plot below, we add a layer for water information and a coord_sf() layer to restrict the x and y-axis limits to the Twin Cities bounding box. (Without this layer, the map would zoom back out to show all counties and bodies of water).
ggplot() +
geom_raster(data = elev_df, aes(x = x, y = y, fill = elevation)) +
geom_sf(data = mn_counties, fill = NA, color = "black") + # county boundary layer
geom_sf(data = mn_water, fill = "lightsteelblue1", color = "lightsteelblue1") + # NEW: river/lake layer
geom_sf(data = mn_cities %>% filter(Population >= 10000), mapping = aes(color = Population, size = Population)) + # cities layer
scale_color_viridis_c(option = "magma") + # continuous (gradient) color scale
scale_fill_gradient(low = "darkgreen", high = "white") + # continuous (gradient) fill scale
coord_sf(xlim = seven_countyarea[c("xmin", "xmax")], ylim = seven_countyarea[c("ymin", "ymax")]) + # NEW: crop map to Twin Cities bounding box
labs(title = "Twin Cities with Population >= 10,000") +
ggthemes::theme_map() +
theme(legend.position = "none") # remove legendLet’s add to the above code chunk to save the map above to an image file called tc_map_zoom.png in the figures folder. The code example below shows a general template for saving a plot to file. Choose a reasonable width and height. (There are also jpeg() and pdf() functions for writing images.)
png("relative path to image", width = width_in_pixels, height = height_in_pixels)
# Code for creating plot
dev.off()
Going Beyond - Twin Cities map with leaflet
Below we show how to make the MN counties map in the leaflet package.
library(leaflet)
mn_counties_leaf <- mn_counties %>% st_transform(4326) # Leaflet expects this CRS for vectors
mn_cities_leaf <- mn_cities %>% st_transform(4326)
cities_per_county <- st_join(mn_cities_leaf, mn_counties_leaf) %>%
st_drop_geometry() %>% # removes geometry - makes the following calculation more efficient
count(name)
mn_counties_leaf %>%
filter(name %in% c("Anoka", "Hennepin", "Ramsey", "Dakota", "Carver", "Washington", "Scott")) %>%
left_join(cities_per_county) %>%
leaflet() %>%
addProviderTiles("CartoDB.Positron") %>%
addPolygons(
color = "#444444", weight = 1, smoothFactor = 0.5, opacity = 1.0,
fillOpacity = 0.5, fillColor = ~colorQuantile("YlOrRd", n)(n),
highlightOptions = highlightOptions(color = "white", weight = 2, bringToFront = TRUE)) %>%
addCircles(data = mn_cities_leaf %>% filter(County %in% paste(c("Anoka", "Hennepin", "Ramsey", "Dakota", "Carver", "Washington", "Scott"), "County")), color = "#444444")Solutions
- Download Shapefiles
- Read in Files
Solution
mn_cities <- st_read("../data/shp_loc_pop_centers/city_and_township_population_centers.shp")Reading layer `city_and_township_population_centers' from data source
`/Users/bheggese/Desktop/Current Teaching/Stat:Comp 212/212_spring_2025/data/shp_loc_pop_centers/city_and_township_population_centers.shp'
using driver `ESRI Shapefile'
Simple feature collection with 1081 features and 8 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: 190832.6 ymin: 4816672 xmax: 747463.4 ymax: 5468045
Projected CRS: NAD83 / UTM zone 15Nmn_water <- st_read("../data/shp_water_lakes_rivers/LakesAndRivers.shp")Reading layer `LakesAndRivers' from data source
`/Users/bheggese/Desktop/Current Teaching/Stat:Comp 212/212_spring_2025/data/shp_water_lakes_rivers/LakesAndRivers.shp'
using driver `ESRI Shapefile'
Simple feature collection with 2313 features and 8 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 419538.6 ymin: 4922700 xmax: 522665 ymax: 5029945
Projected CRS: NAD83 / UTM zone 15N- Change CRS
Solution
nc <- st_read(system.file("shape/nc.shp", package = "sf"))Reading layer `nc' from data source
`/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/sf/shape/nc.shp'
using driver `ESRI Shapefile'
Simple feature collection with 100 features and 14 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
Geodetic CRS: NAD27transform_and_plot <- function(spatial_obj, new_crs, title) {
spatial_obj %>%
st_transform(crs = new_crs) %>%
ggplot() +
geom_sf() +
labs(title = title) +
theme_classic()
}
ggplot(nc) +
geom_sf() +
theme_classic() +
labs(title = "NAD27 (Original)")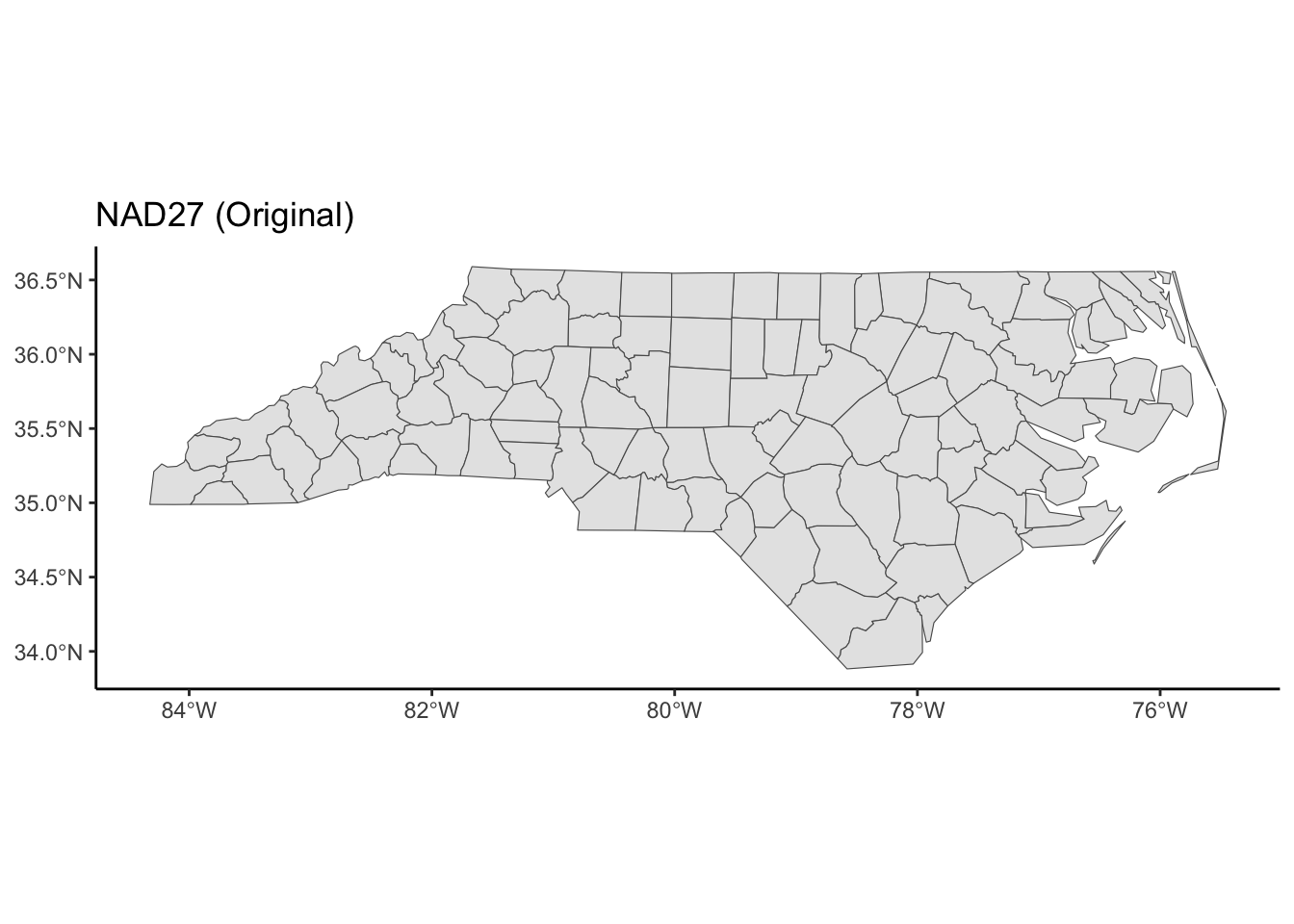
transform_and_plot(nc, new_crs = "EPSG:32133", title = "South Carolina")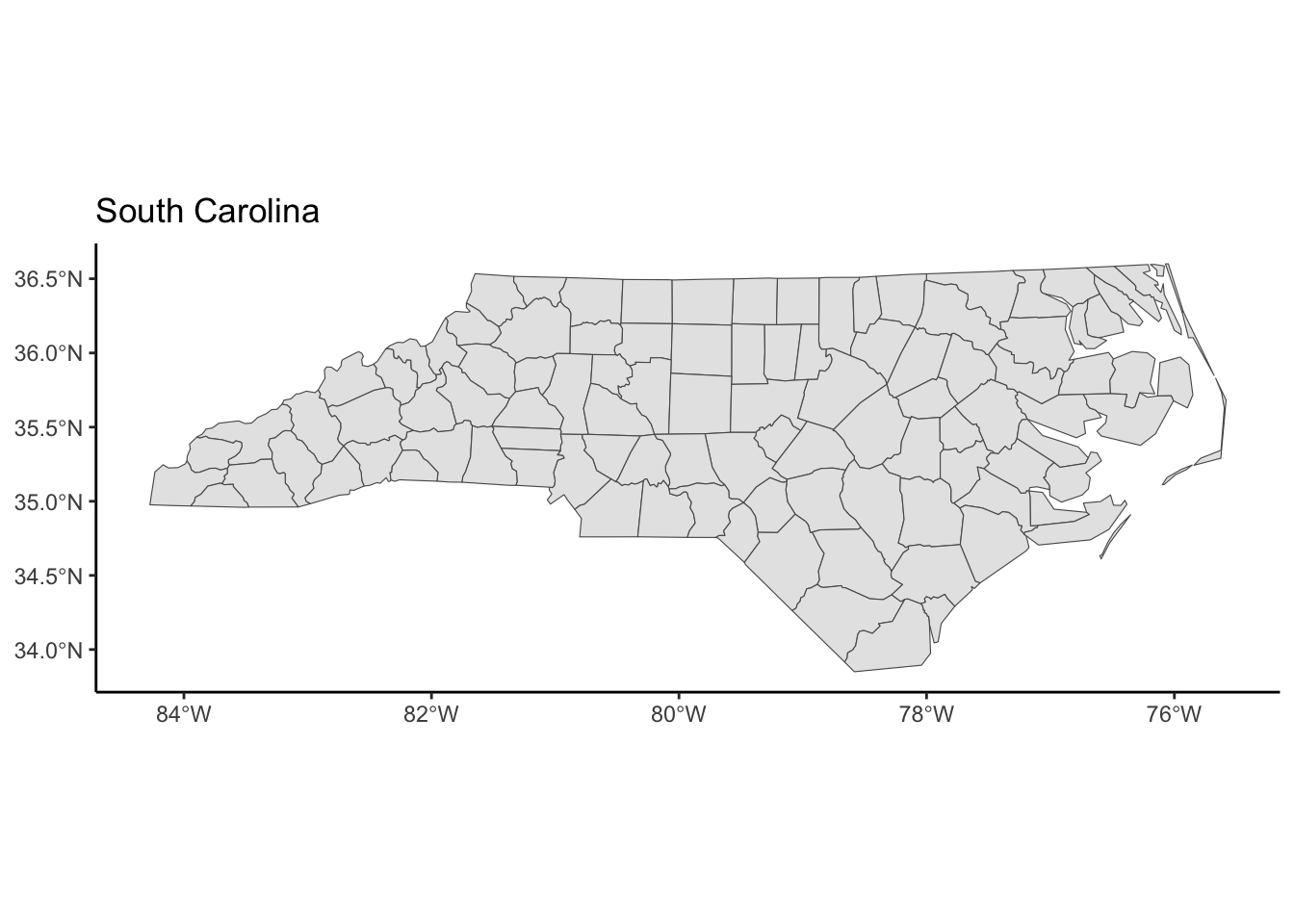
transform_and_plot(nc, new_crs = "EPSG:6439", title = "Florida")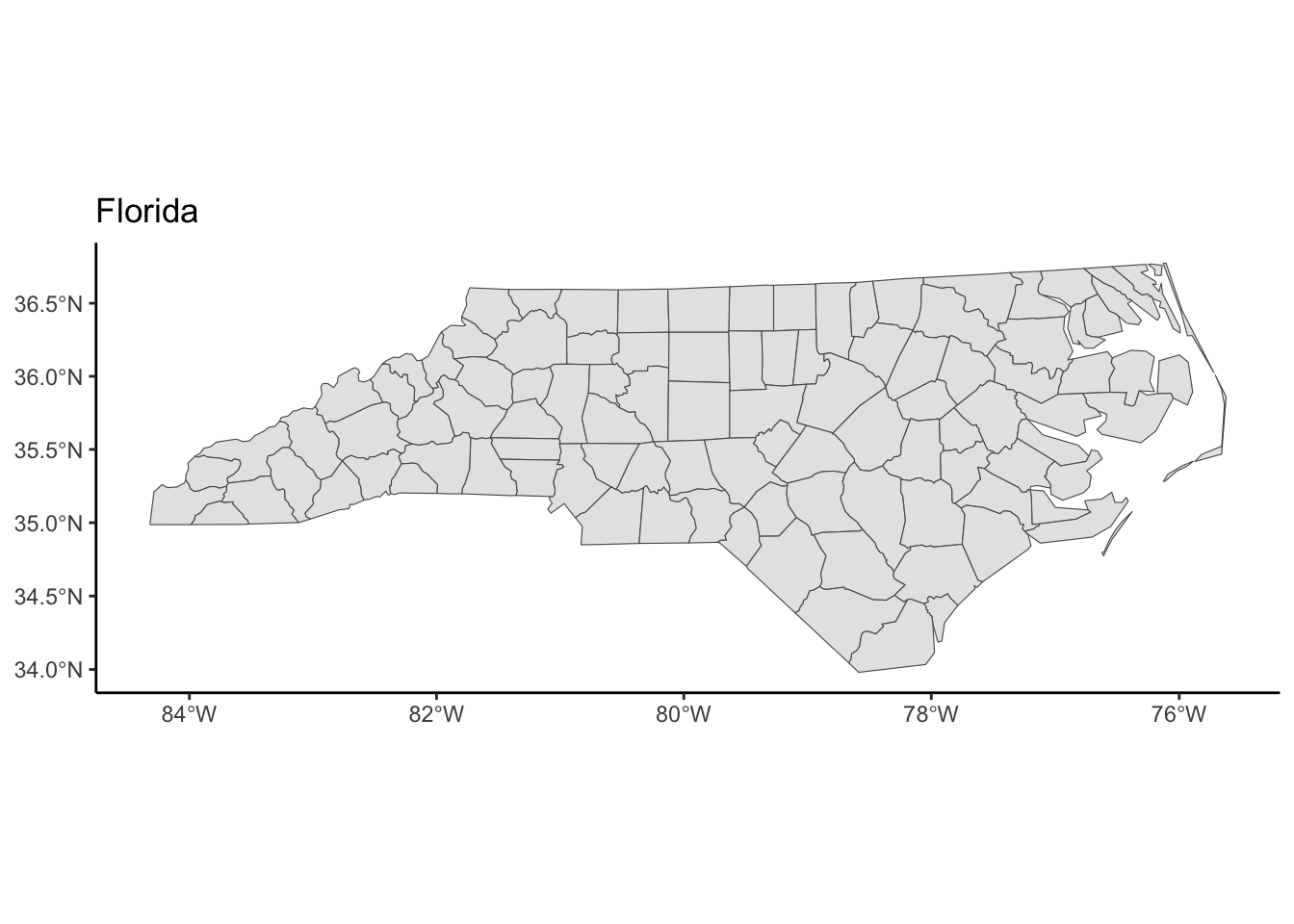
transform_and_plot(nc, new_crs = "EPSG:3112", title = "Australia (GDA94)")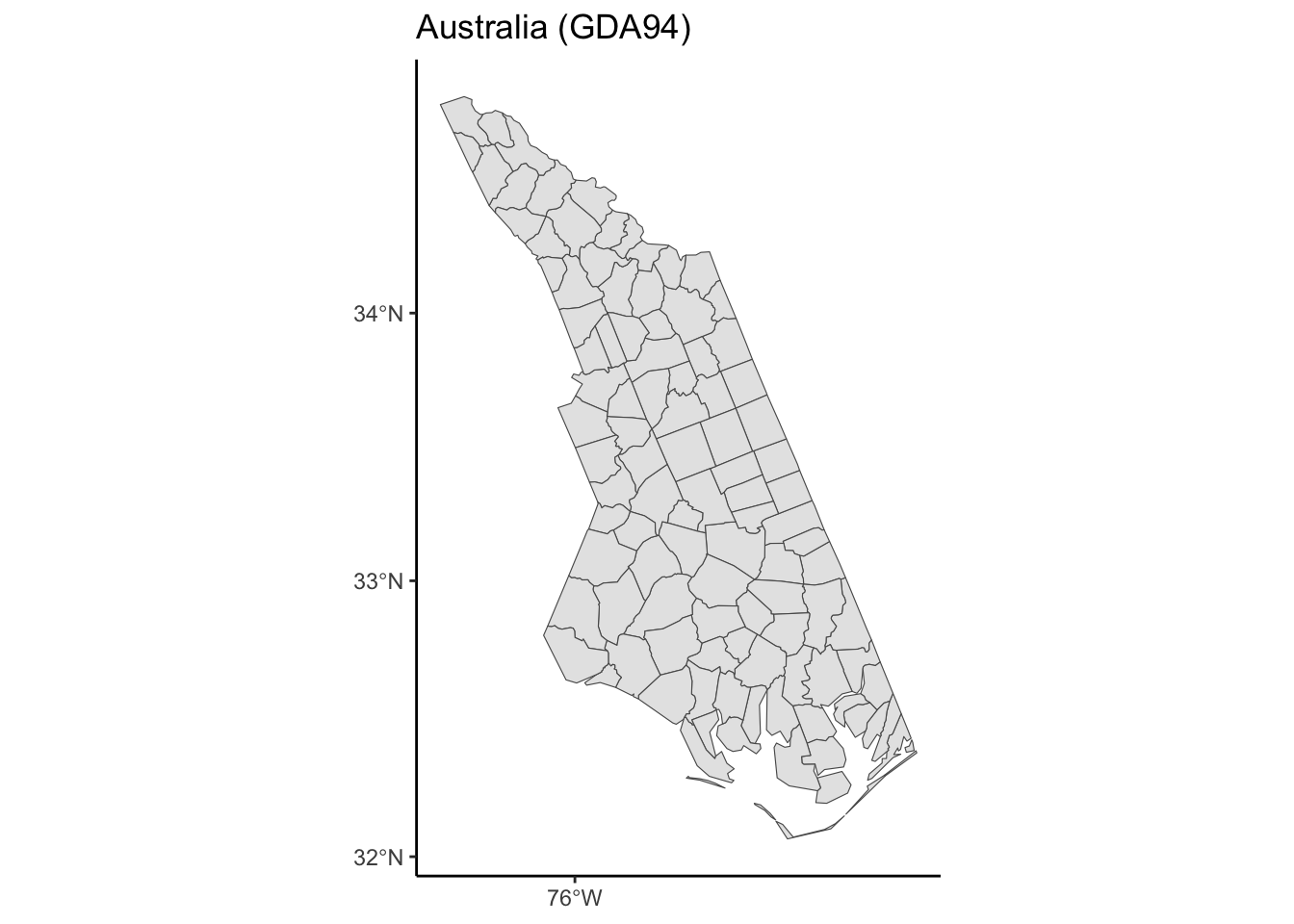
transform_and_plot(nc, new_crs = "EPSG:20353", title = "Australia (Queensland, South, West)")
transform_and_plot(nc, new_crs = "EPSG:5940", title = "Russia")
Get county boundaries
Unifying CRSs across different spatial datasets
Solution
# Check CRSs
st_crs(mn_cities)Coordinate Reference System:
User input: NAD83 / UTM zone 15N
wkt:
PROJCRS["NAD83 / UTM zone 15N",
BASEGEOGCRS["NAD83",
DATUM["North American Datum 1983",
ELLIPSOID["GRS 1980",6378137,298.257222101,
LENGTHUNIT["metre",1]]],
PRIMEM["Greenwich",0,
ANGLEUNIT["degree",0.0174532925199433]],
ID["EPSG",4269]],
CONVERSION["UTM zone 15N",
METHOD["Transverse Mercator",
ID["EPSG",9807]],
PARAMETER["Latitude of natural origin",0,
ANGLEUNIT["Degree",0.0174532925199433],
ID["EPSG",8801]],
PARAMETER["Longitude of natural origin",-93,
ANGLEUNIT["Degree",0.0174532925199433],
ID["EPSG",8802]],
PARAMETER["Scale factor at natural origin",0.9996,
SCALEUNIT["unity",1],
ID["EPSG",8805]],
PARAMETER["False easting",500000,
LENGTHUNIT["metre",1],
ID["EPSG",8806]],
PARAMETER["False northing",0,
LENGTHUNIT["metre",1],
ID["EPSG",8807]]],
CS[Cartesian,2],
AXIS["(E)",east,
ORDER[1],
LENGTHUNIT["metre",1]],
AXIS["(N)",north,
ORDER[2],
LENGTHUNIT["metre",1]],
ID["EPSG",26915]]st_crs(mn_water)Coordinate Reference System:
User input: NAD83 / UTM zone 15N
wkt:
PROJCRS["NAD83 / UTM zone 15N",
BASEGEOGCRS["NAD83",
DATUM["North American Datum 1983",
ELLIPSOID["GRS 1980",6378137,298.257222101,
LENGTHUNIT["metre",1]]],
PRIMEM["Greenwich",0,
ANGLEUNIT["degree",0.0174532925199433]],
ID["EPSG",4269]],
CONVERSION["UTM zone 15N",
METHOD["Transverse Mercator",
ID["EPSG",9807]],
PARAMETER["Latitude of natural origin",0,
ANGLEUNIT["Degree",0.0174532925199433],
ID["EPSG",8801]],
PARAMETER["Longitude of natural origin",-93,
ANGLEUNIT["Degree",0.0174532925199433],
ID["EPSG",8802]],
PARAMETER["Scale factor at natural origin",0.9996,
SCALEUNIT["unity",1],
ID["EPSG",8805]],
PARAMETER["False easting",500000,
LENGTHUNIT["metre",1],
ID["EPSG",8806]],
PARAMETER["False northing",0,
LENGTHUNIT["metre",1],
ID["EPSG",8807]]],
CS[Cartesian,2],
AXIS["(E)",east,
ORDER[1],
LENGTHUNIT["metre",1]],
AXIS["(N)",north,
ORDER[2],
LENGTHUNIT["metre",1]],
ID["EPSG",26915]]st_crs(mn_counties) # mn_counties is different!Coordinate Reference System:
User input: EPSG:4326
wkt:
GEOGCRS["WGS 84",
DATUM["World Geodetic System 1984",
ELLIPSOID["WGS 84",6378137,298.257223563,
LENGTHUNIT["metre",1]]],
PRIMEM["Greenwich",0,
ANGLEUNIT["degree",0.0174532925199433]],
CS[ellipsoidal,2],
AXIS["geodetic latitude (Lat)",north,
ORDER[1],
ANGLEUNIT["degree",0.0174532925199433]],
AXIS["geodetic longitude (Lon)",east,
ORDER[2],
ANGLEUNIT["degree",0.0174532925199433]],
USAGE[
SCOPE["Horizontal component of 3D system."],
AREA["World."],
BBOX[-90,-180,90,180]],
ID["EPSG",4326]]# Transform the CRS of county data to the more local CRS of the cities
mn_counties <- mn_counties %>%
st_transform(crs = st_crs(mn_cities))
# Check the new CRS for mn_counties
st_crs(mn_counties)Coordinate Reference System:
User input: NAD83 / UTM zone 15N
wkt:
PROJCRS["NAD83 / UTM zone 15N",
BASEGEOGCRS["NAD83",
DATUM["North American Datum 1983",
ELLIPSOID["GRS 1980",6378137,298.257222101,
LENGTHUNIT["metre",1]]],
PRIMEM["Greenwich",0,
ANGLEUNIT["degree",0.0174532925199433]],
ID["EPSG",4269]],
CONVERSION["UTM zone 15N",
METHOD["Transverse Mercator",
ID["EPSG",9807]],
PARAMETER["Latitude of natural origin",0,
ANGLEUNIT["Degree",0.0174532925199433],
ID["EPSG",8801]],
PARAMETER["Longitude of natural origin",-93,
ANGLEUNIT["Degree",0.0174532925199433],
ID["EPSG",8802]],
PARAMETER["Scale factor at natural origin",0.9996,
SCALEUNIT["unity",1],
ID["EPSG",8805]],
PARAMETER["False easting",500000,
LENGTHUNIT["metre",1],
ID["EPSG",8806]],
PARAMETER["False northing",0,
LENGTHUNIT["metre",1],
ID["EPSG",8807]]],
CS[Cartesian,2],
AXIS["(E)",east,
ORDER[1],
LENGTHUNIT["metre",1]],
AXIS["(N)",north,
ORDER[2],
LENGTHUNIT["metre",1]],
ID["EPSG",26915]]- Counties and cities
Solution
# Option 1
ggplot() + # plot frame
geom_sf(data = mn_counties, fill = "white") + # county boundary layer
geom_sf(data = mn_cities, size = 0.5) + # city point layer
ggthemes::theme_map()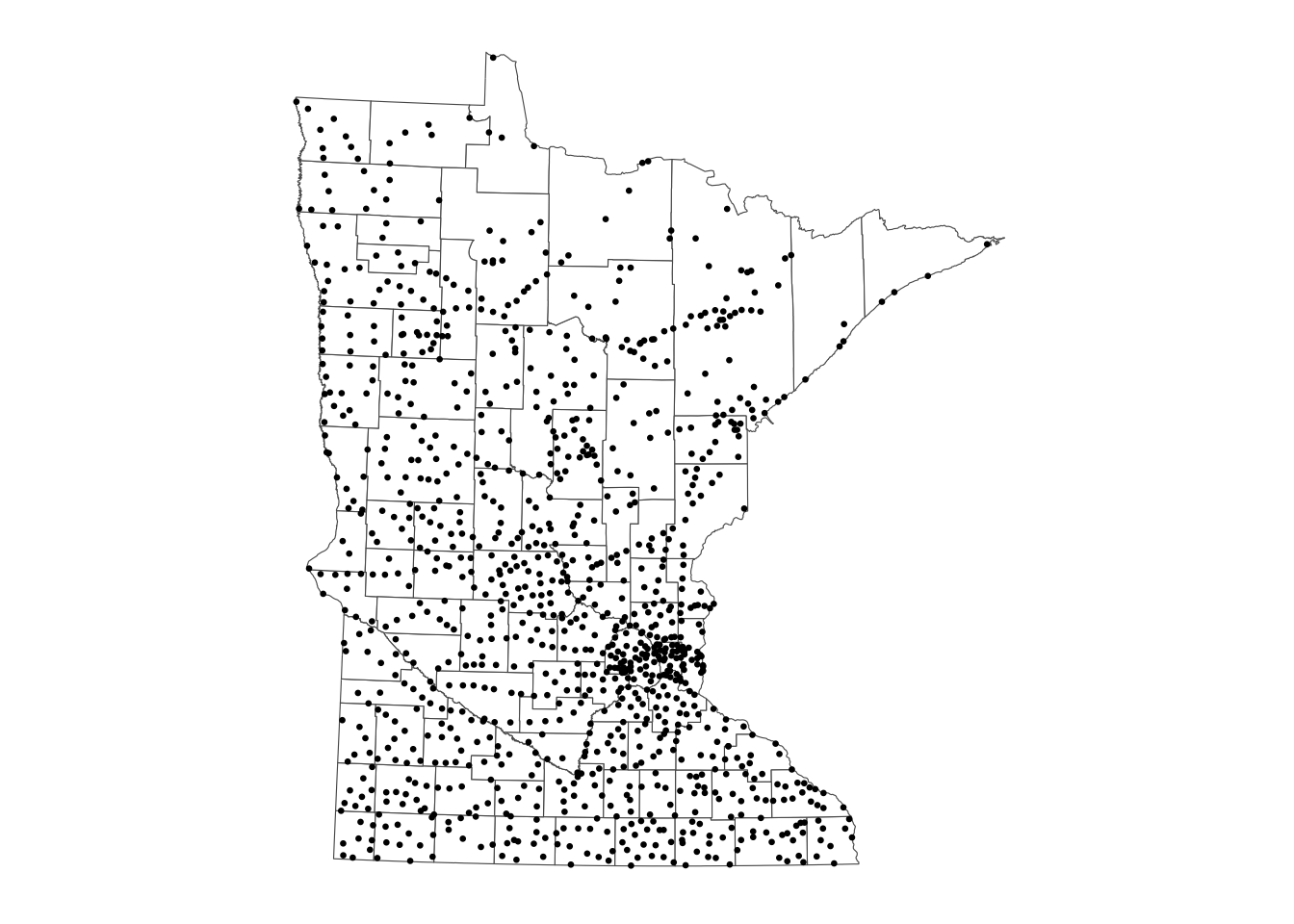
# Option 2
ggplot(mn_counties) + # plot frame
geom_sf(fill = "white") + # county boundary layer
geom_sf(data = mn_cities, size = 0.5) + # city point layer
ggthemes::theme_map()
- Customize Colors
Solution
The viridis color scale results in plots that can be interpreted analogously whether in color or black and white and is color-blind friendly.
- The
_d()variant is used when color is mapped to a discrete (categorical) variable. - The
_c()variant is used when color is mapped to a continuous variable. - The
_b()variant is used when color is mapped to a continuous variable but when we want that continuous variable to be binned so that there is a small set of colors.
- Adding elevation raster data
Solution
ggplot() +
geom_raster(data = elev_df, aes(x = x, y = y, fill = elevation)) + # adding the elevation as first (bottom) layer
scale_fill_gradient(low = "darkgreen", high = "white", guide = FALSE) +
geom_sf(data = mn_counties, fill = NA, color = "black") +
geom_sf(data = mn_cities %>% filter(Population >= 10000), mapping = aes(color = Population, size = Population))+ # cities layer
scale_color_viridis_c() + # continuous (gradient) color scale
labs(title = "Minnesota Cities with Population >= 10,000") +
ggthemes::theme_map() +
theme(legend.position = "bottom") # move legend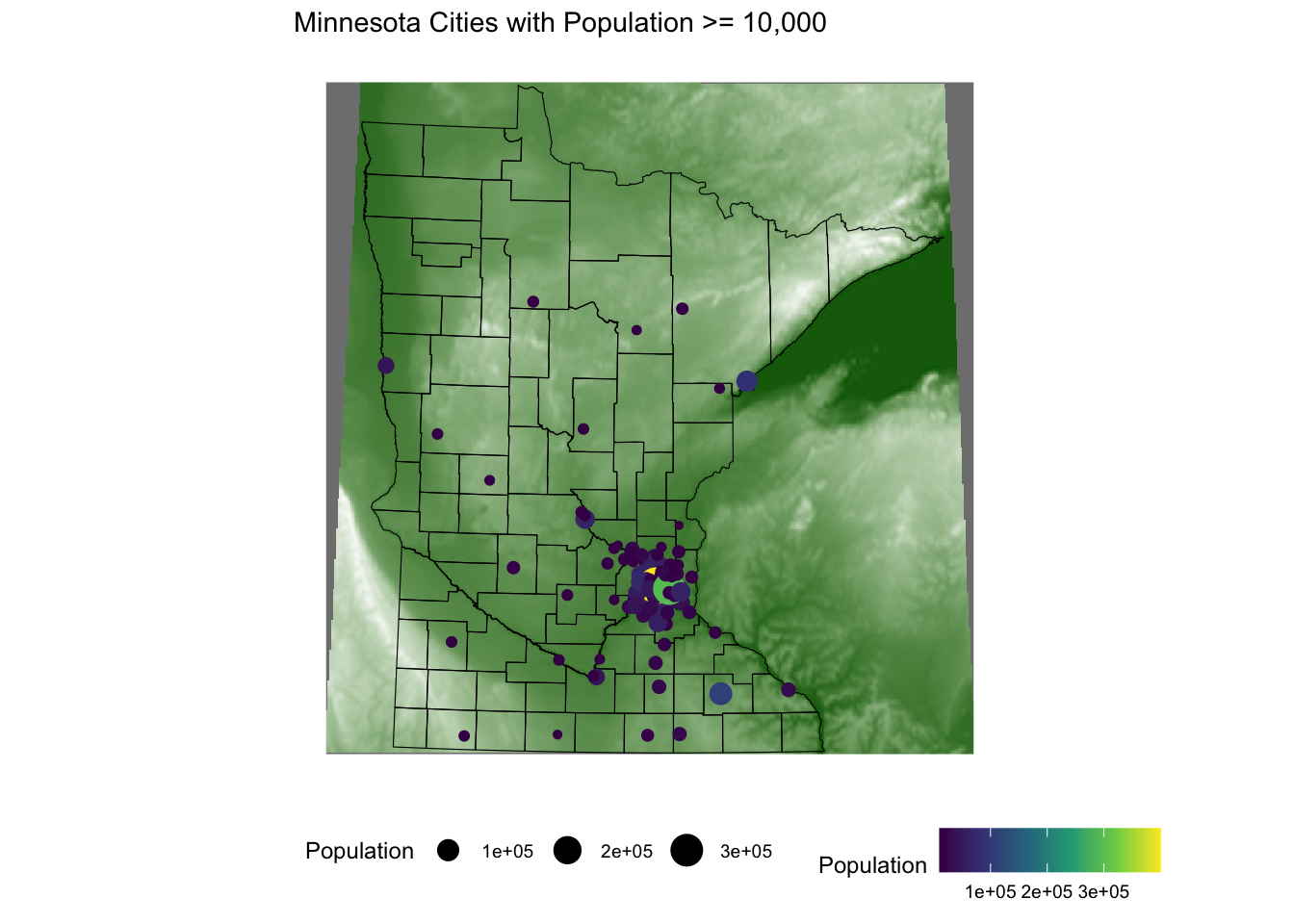
- Zoom in to Twin Cities and add water
Solution
seven_countyarea <- mn_counties %>%
filter(name %in% c("Anoka", "Hennepin", "Ramsey", "Dakota", "Carver", "Washington", "Scott")) %>%
st_bbox()
seven_countyarea xmin ymin xmax ymax
419967.1 4924212.8 521254.8 5029157.4 #sf version: 1.0-19
#elevatr version: 0.99.0
elevation <- elevatr::get_elev_raster(mn_counties %>% st_crop(seven_countyarea), z = 9, clip = "bbox")
raster::crs(elevation) <- sf::st_crs(mn_counties)
# Convert to data frame for plotting
elev_df <- elevation %>% terra::as.data.frame(xy = TRUE)
colnames(elev_df) <- c("x", "y", "elevation")png("figures/tc_map_zoom.png", width = 800, height = 400)
ggplot() +
geom_raster(data = elev_df, aes(x = x, y = y, fill = elevation)) +
geom_sf(data = mn_counties, fill = NA, color = "black") + # county boundary layer
geom_sf(data = mn_water, fill = "lightsteelblue1", color = "lightsteelblue1") + # NEW: river/lake layer
geom_sf(data = mn_cities %>% filter(Population >= 10000), mapping = aes(color = Population, size = Population)) + # cities layer
scale_color_viridis_c(option = "magma") + # continuous (gradient) color scale
scale_fill_gradient(low = "darkgreen", high = "white") + # continuous (gradient) fill scale
coord_sf(xlim = seven_countyarea[c("xmin", "xmax")], ylim = seven_countyarea[c("ymin", "ymax")]) + # NEW: crop map to Twin Cities bounding box
labs(title = "Twin Cities with Population >= 10,000") +
ggthemes::theme_map() +
theme(legend.position = "none") # remove legend
dev.off()Homework 2
Navigate to Homework 2 and find the two parts. This homework involves reviewing ggplot2 tools to replicate two separate visualizations.
Homework 3
Navigate to Homework 3 and find the GitHub Classroom to clone the repository for this homework. These two parts involves using the tools we learned today to spatially explore different aspects of the Twin Cities.
Reflect
Consider the following:
- What was challenging and why? What specific strategy do you want to try to improve your understanding?
- What strategies, associations, insights helped things click today?
After Class
- Start thinking concretely about your course project topic. Project Milestone 1 is coming up.
- Start looking for datasets to see what might be feasible. Think about who you would like to work with.
- If you are looking for project partners, there will be time on next class to connect with partners.
- Take a look at the Schedule page to see how to prepare for the next class.
- Work on Homework 2. Due next Tuesday.
- Start on Homework 3. Due the following Tuesday.