Topic 4 Cross-validation
Learning Goals
- Accurately describe all steps of cross-validation to estimate the test/out-of-sample version of a model evaluation metric
- Explain what role CV has in a predictive modeling analysis and its connection to overfitting
- Explain the pros/cons of higher vs. lower k in k-fold CV in terms of sample size and computing time
- Implement cross-validation in R using the
tidymodelspackage - Using these tools and concepts to:
- Inform and justify data analysis and modeling process and the resulting conclusions with clear, organized, logical, and compelling details that adapt to the background, values, and motivations of the audience and context in which communication occurs
Notes: k-fold Cross-Validation
CONTEXT
world = supervised learning
We want to build a model some output variable \(y\) by some predictors \(x\).task = regression
\(y\) is quantitativemodel = linear regression model via least squares algorithm
We’ll assume that the relationship between \(y\) and \(x\) can be represented by\[y = \beta_0 + \beta_1 x_1 + \beta_2 x_2 + ... + \beta_p x_p + \varepsilon\]
GOAL: model evaluation
We want more honest metrics of prediction quality that
- assess how well our model predicts new outcomes; and
- help prevent overfitting.

Why is overfitting so bad?
Not only can overfitting produce misleading models, it can have serious societal impacts. Examples:
A former Amazon algorithm built to help sift through resumes was overfit to its current employees in leadership positions (who weren’t representative of the general population or candidate pool).
Facial recognition algorithms are often overfit to the people who build them (who are not broadly representative of society). As one example, this has led to disproportionate bias in policing. For more on this topic, you might check out Coded Bias, a documentary by Shalini Kantayya which features MIT Media Lab researcher Joy Buolamwini.
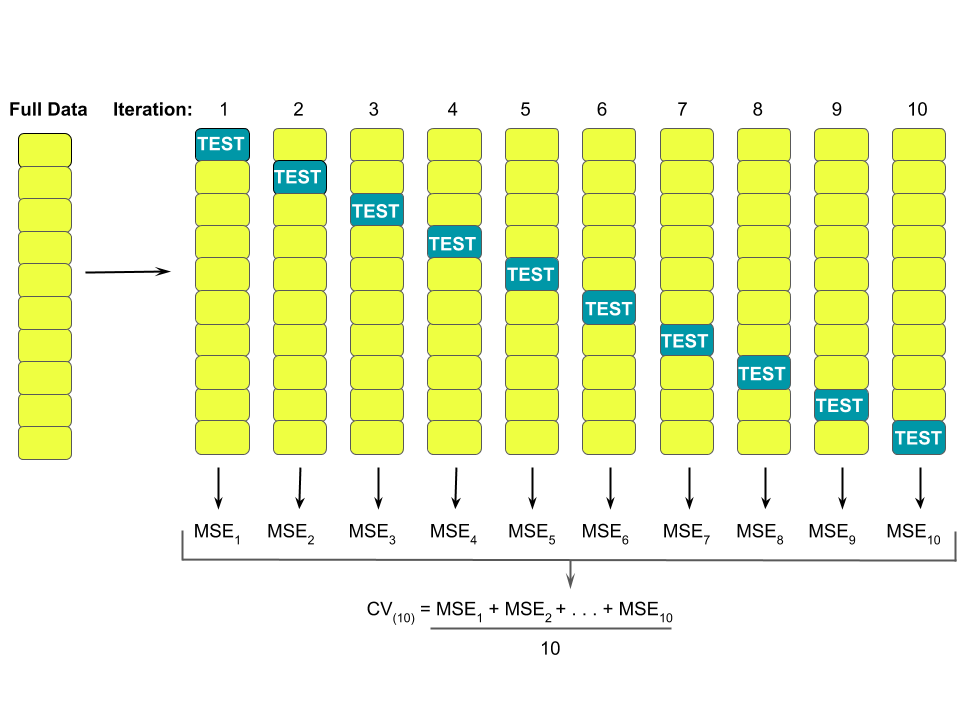
We can use k-fold cross-validation to estimate the typical error in our model predictions for new data:
- Divide the data into \(k\) folds (or groups) of approximately equal size.
- Repeat the following procedures for each fold \(j = 1,2,...,k\):
- Remove fold \(j\) from the data set.
- Fit a model using the data in the other \(k-1\) folds (training).
- Use this model to predict the responses for the \(n_j\) cases in fold \(j\): \(\hat{y}_1, ..., \hat{y}_{n_j}\).
- Calculate the MAE for fold \(j\) (testing): \(\text{MAE}_j = \frac{1}{n_j}\sum_{i=1}^{n_j} |y_i - \hat{y}_i|\).
- Remove fold \(j\) from the data set.
- Combine this information into one measure of model quality: \[\text{CV}_{(k)} = \frac{1}{k} \sum_{j=1}^k \text{MAE}_j\]
Small Group Discussion: Algorithms and Tuning
Definitions:
algorithm = a step-by-step procedure for solving a problem (Merriam-Webster)
tuning parameter = a parameter or quantity upon which an algorithm depends, that must be selected or tuned to “optimize” the algorithm
Prompts:
- Algorithms
Why is \(k\)-fold cross-validation an algorithm?
What is the tuning parameter of this algorithm and what values can this take?
Solution
- Yes. It follows a list of steps to get to its goal.
- \(k\), the number of folds, is a tuning parameter. \(k\) can be any integer from 1, 2, …, \(n\) where \(n\) is our sample size.
- Tuning the k-fold Cross-Validation algorithm
Let’s explore k-fold cross-validation with some personal experience. Our class has a representative sample of cards from a non-traditional population (no “face cards”, not equal numbers, etc). We want to use these to predict whether a new card will be odd or even (a classification task).
- Based on all of our cards, do we predict the next card will be odd or even?
- You’ve been split into 2 groups. Use 2-fold cross-validation to estimate the possible error of using our sample of cards to predict whether a new card will be odd or even. How’s this different than validation?
- Repeat for 3-fold cross-validation. Why might this be better than 2-fold cross-validation?
- Repeat for LOOCV, i.e. n-fold cross-validation where n is the number of students in this room. Why might this be worse than 3-fold cross-validation?
- What value of k do you think practitioners typically use?
Solution
- Use the percentage of odd and percentage of even among the sample of cards to help you make a prediction.
- We use both groups as training and testing, in turn.
- We have a larger dataset to train our model on. We are less likely to get an unrepresentative set as our training data.
- Prediction error for 1 person is highly variable.
- In practice, \(k = 10\) and \(k=7\) are common choices for cross-validation. This has been shown to hit the ‘sweet spot’ between the extremes of \(k=n\) (LOOCV) and \(k=2\).
- \(k=2\) only utilizes 50% of the data for each training model, thus might result in overestimating the prediction error
- \(k=n\) leave-one-out cross-validation (LOOCV) requires us to build \(n\) training models, thus might be computationally expensive for larger sample sizes \(n\). Further, with only one data point in each test set, the training sets have a lot of overlap. This correlation among the training sets can make the ultimate corresponding estimate of prediction error less reliable.
- R Code Preview
We’ve been doing a 2-step process to build linear regression models using the tidymodels package:
# STEP 1: model specification
lm_spec <- linear_reg() %>%
set_mode("regression") %>%
set_engine("lm")
# STEP 2: model estimation
my_model <- lm_spec %>%
fit(
y ~ x1 + x2,
data = sample_data
)For k-fold cross-validation, we can tweak STEP 2.
- Discuss the code below and why we need to set the seed.
# k-fold cross-validation
set.seed(___)
my_model_cv <- lm_spec %>%
fit_resamples(
y ~ x1 + x2,
resamples = vfold_cv(sample_data, v = ___),
metrics = metric_set(mae, rsq)
)Solution
The process of creating the folds is random, so we should set the seed to have reproducibility within our work.Notes: R code
Suppose we wish to build and evaluate a linear regression model of y vs x1 and x2 using our sample_data.
Obtain k-fold cross-validated estimates of MAE and \(R^2\)
(Review above for discussion of these steps.)
# model specification
lm_spec <- linear_reg() %>%
set_mode("regression") %>%
set_engine("lm")
# k-fold cross-validation
# For "v", put your number of folds k
set.seed(___)
model_cv <- lm_spec %>%
fit_resamples(
y ~ x1 + x2,
resamples = vfold_cv(sample_data, v = ___),
metrics = metric_set(mae, rsq)
)
Obtain the cross-validated metrics
Details: get the MAE and R-squared for each test fold
Exercises
# Load packages and data
library(tidyverse)
library(tidymodels)
humans <- read.csv("https://bcheggeseth.github.io/253_spring_2024/data/bodyfat50.csv") %>%
filter(ankle < 30) %>%
rename(body_fat = fatSiri)
Review: In-sample metrics
Use the
humansdata to build two separate models ofheight:
# STEP 2: model estimation
model_1 <- ___ %>%
___(height ~ hip + weight + thigh + knee + ankle, data = humans)
model_2 <- ___ %>%
___(height ~ chest * age * weight * body_fat + abdomen + hip + thigh + knee + ankle + biceps + forearm + wrist, data = humans)Calculate the **in-sample** R-squared for both models:Calculate the **in-sample** MAE for both models:# IN-SAMPLE MAE for model_1 = ???
model_1 %>%
___(new_data = ___) %>%
mae(truth = ___, estimate = ___)# IN-SAMPLE MAE for model_2 = ???
model_2 %>%
___(new_data = ___) %>%
mae(truth = ___, estimate = ___)Solution
# STEP 1: model specification
lm_spec <- linear_reg() %>%
set_mode("regression") %>%
set_engine("lm")
# STEP 2: model estimation
model_1 <- lm_spec %>%
fit(height ~ hip + weight + thigh + knee + ankle, data = humans)
model_2 <- lm_spec %>%
fit(height ~ chest * age * weight * body_fat + abdomen + hip + thigh + knee + ankle + biceps + forearm + wrist, data = humans)
# IN-SAMPLE R^2 for model_1 = 0.40
model_1 %>%
glance()
# IN-SAMPLE R^2 for model_2 = 0.87
model_2 %>%
glance()
# IN-SAMPLE MAE for model_1 = 1.55
model_1 %>%
augment(new_data = humans) %>%
mae(truth = height, estimate = .pred)
# IN-SAMPLE MAE for model_2 = 0.64
model_2 %>%
augment(new_data = humans) %>%
mae(truth = height, estimate = .pred)- In-sample model comparison
Which model seems “better” by the in-sample metrics you calculated above? Any concerns about either of these models?
Solution
The in-sample metrics are better formodel_2, but from experience in our previous class, we should expect this to be overfit.
10-fold CV
Complete the code to run 10-fold cross-validation for our two models.model_1:height ~ hip + weight
model_2:height ~ chest * age * weight * body_fat + abdomen + hip + thigh + knee + ankle + biceps + forearm + wrist
# 10-fold cross-validation for model_1
set.seed(253)
model_1_cv <- ___ %>%
___(
___,
___ = vfold_cv(___, v = ___),
___ = metric_set(mae, rsq)
)# 10-fold cross-validation for model_2
set.seed(253)
model_2_cv <- ___ %>%
___(
___,
___ = vfold_cv(___, v = ___),
___ = metric_set(mae, rsq)
)Solution
# 10-fold cross-validation for model_1
set.seed(253)
model_1_cv <- lm_spec %>%
fit_resamples(
height ~ hip + weight + thigh + knee + ankle,
resamples = vfold_cv(humans, v = 10),
metrics = metric_set(mae, rsq)
)
# STEP 2: 10-fold cross-validation for model_2
set.seed(253)
model_2_cv <- lm_spec %>%
fit_resamples(
height ~ chest * age * weight * body_fat + abdomen + hip + thigh + knee + ankle + biceps + forearm + wrist,
resamples = vfold_cv(humans, v = 10),
metrics = metric_set(mae, rsq)
)- Calculating the CV MAE
- Use
collect_metrics()to obtain the cross-validated MAE and \(R^2\) for both models.
- Interpret the cross-validated MAE and \(R^2\) for
model_1.
Solution
# model_1
# CV MAE = 1.87, CV R-squared = 0.40
model_1_cv %>%
collect_metrics()
# model_2
# CV MAE = 2.47, CV R-squared = 0.52
model_2_cv %>%
collect_metrics()- We expect our first model to explain roughly 40% of variability in height among new adults, and to produce predictions of height that are off by 1.9 inches on average.
Details: fold-by-fold results
collect_metrics()gave the final CV MAE, or the average MAE across all 10 test folds.unnest(.metrics)provides the MAE from each test fold.Obtain the fold-by-fold results for the
model_1cross-validation procedure usingunnest(.metrics).Which fold had the worst average prediction error and what was it?
Recall that
collect_metrics()reported a final CV MAE of 1.87 formodel_1. Confirm this calculation by wrangling the fold-by-fold results from part a.
Solution
Comparing models
The table below summarizes the in-sample and 10-fold CV MAE for both models.
Model IN-SAMPLE MAE 10-fold CV MAE model_11.55 1.87 model_20.64 2.47
- Based on the in-sample MAE alone, which model appears better?
- Based on the CV MAE alone, which model appears better?
- Based on all of these results, which model would you pick?
- Do the in-sample and CV MAE suggest that
model_1is overfit to ourhumanssample data? What aboutmodel_2?
- Based on the in-sample MAE alone, which model appears better?
Solution
model_2model_1model_1–model_2produces bad predictions for new adultsmodel_1is NOT overfit – its predictions of height for new adults seem roughly as accurate as the predictions for the adults in our sample.model_2IS overfit – its predictions of height for new adults are worse than the predictions for the adults in our sample.
- LOOCV
- Reconsider
model_1. Instead of estimating its prediction accuracy using the 10-fold CV MAE, use the LOOCV MAE. THINK: How many people are in ourhumanssample? - How does the LOOCV MAE compare to the 10-fold CV MAE of 1.87? NOTE: These are just two different approaches to estimating the same thing: the typical prediction error when applying our model to new data. Thus we should expect them to be similar.
- Explain why we technically don’t need to
set.seed()for the LOOCV algorithm.
- Reconsider
Solution
- There are 40 people in our sample, thus LOOCV is equivalent to 40-fold CV:
nrow(humans)
model_1_loocv <- lm_spec %>%
fit_resamples(
height ~ hip + weight + thigh + knee + ankle,
resamples = vfold_cv(humans, v = nrow(humans)),
metrics = metric_set(mae)
)
model_1_loocv %>%
collect_metrics()- The LOOCV MAE (1.82) is very similar to the 10-fold CV MAE (1.96).
- There’s no randomness in the test folds. Each test fold is a single person.
- Data drill
- Calculate the average height of people under 40 years old vs people 40+ years old.
- Plot height vs age among our subjects that are 30+ years old.
- Fix this code:
Solution
- Reflection: Part 1
The “regular” exercises are over but class is not done! Your group should agree to either work on HW2 OR the remaining reflection questions.
This is the end of Unit 1 on “Regression: Model Evaluation”! Let’s reflect on the technical content of this unit:
- What was the main motivation / goal behind this unit?
- What are the four main questions that were important to this unit?
- For each of the following tools, describe how they work and what questions they help us address:
- R-squared
- residual plots
- out-of-sample MAE
- in-sample MAE
- validation
- cross-validation
- In your own words, define the following: overfitting, algorithm, tuning parameter.
- Review the new tidymodels syntax from this unit. Identify key themes and patterns.
I encourage you to make a copy of this document and add notes/thoughts after this Unit 1.
Reflection: Part 2
The reflection above addresses your learning goals in this course. Consider the other components that have helped you worked toward this learning throughout Unit 1.With respect to collaboration, reflect upon your strengths and what you might change in the next unit:
- How actively did you contribute to group discussions?
- How actively did you include all other group members in discussion?
- In what ways did you (or did you not) help create a space where others feel comfortable making mistakes & sharing their ideas?
With respect to engagement, reflect upon your strengths and what you might change the next unit:
- Did you regularly attend, be on time for, & stay for the full class sessions?
- Have you not missed more than 3 in-person class sessions?
- Were you actively present during class (eg: not on your phone, not working on other courses, etc)?
- Did you stay updated on Slack?
- When you had questions, did you ask them on Slack or in OH?
With respect to preparation, how many of checkpoints 1–3 did you complete and pass?
With respect to exploration, did you complete and pass HW1? Are you on track to complete and pass HW2?
Done!
- Knit your notes.
- Check the solutions in the online manual.
- Check out the wrap-up steps below.
- If you finish all that during class, work on Homework 2!

