Topic 11 Logistic Regression: Model Building & Evaluation
Learning Goals
- Use a logistic regression model to make hard (class) and soft (probability) predictions
- Interpret non-intercept coefficients from logistic regression models in the data context
Notes: Logistic Regression
CONTEXT
world = supervised learning
We want to model some output variable \(y\) using a set of potential predictors (\(x_1, x_2, ..., x_p\)).task = CLASSIFICATION
\(y\) is categorical and binary(parametric) algorithm
logistic regression
GOAL
Use the parametric logistic regression model to model and classify \(y\).
Logistic Regression
Let \(y\) be a binary categorical response variable:
\[y = \begin{cases} 1 & \; \text{ if event happens} \\ 0 & \; \text{ if event doesn't happen} \\ \end{cases}\]Further define \[\begin{split} p &= \text{ probability event happens} \\ 1-p &= \text{ probability event doesn't happen} \\ \text{odds} & = \text{ odds event happens} = \frac{p}{1-p} \\ \end{split}\]
Then a logistic regression model of \(y\) by \(x\) is \[\begin{split} \log(\text{odds}) & = \beta_0 + \beta_1 x \\ \text{odds} & = e^{\beta_0 + \beta_1 x} \\ p & = \frac{\text{odds}}{\text{odds}+1} = \frac{e^{\beta_0 + \beta_1 x}}{e^{\beta_0 + \beta_1 x}+1} \\ \end{split}\]
Coefficient interpretation
\[\begin{split} \beta_0 & = \text{ LOG(ODDS) when } x=0 \\ e^{\beta_0} & = \text{ ODDS when } x=0 \\ \beta_1 & = \text{ unit change in LOG(ODDS) per 1 unit increase in } x \\ e^{\beta_1} & = \text{ multiplicative change in ODDS per 1 unit increase in } x \\ \end{split}\]
EXAMPLE 1: Check out the data
# Load packages
library(tidyverse)
library(tidymodels)
# Load weather data from rattle package
library(rattle)
data("weatherAUS")
# Wrangle data
sydney <- weatherAUS %>%
filter(Location == "Sydney") %>%
select(RainTomorrow, Humidity9am, Sunshine)
# Check it out
head(sydney)## # A tibble: 6 × 3
## RainTomorrow Humidity9am Sunshine
## <fct> <int> <dbl>
## 1 Yes 92 0
## 2 Yes 83 2.7
## 3 Yes 88 0.1
## 4 Yes 83 0
## 5 Yes 88 0
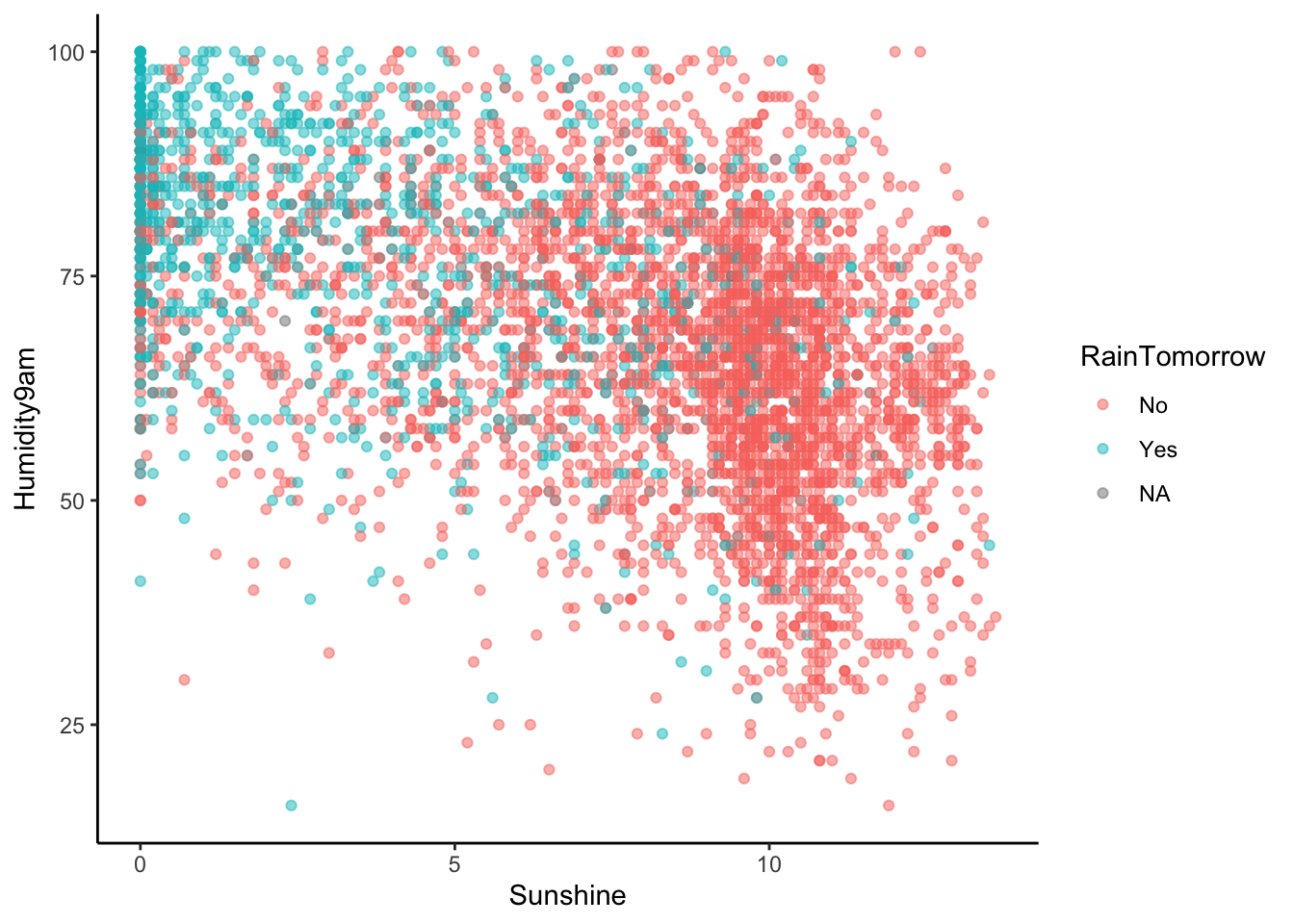
## 6 Yes 69 8.6Let’s model RainTomorrow, whether or not it rains tomorrow in Sydney, by two predictors:
Humidity9am(% humidity at 9am today)Sunshine(number of hours of bright sunshine today)
Check out & comment on the relationship of rain with these 2 predictors:
# Store so we can modify later
rain_plot <- ggplot(sydney, aes(y = Humidity9am, x = Sunshine, color = RainTomorrow)) +
geom_point(alpha = 0.5)EXAMPLE 2: Interpreting coefficients
The logistic regression model is:
- log(odds of rain) = -1.01 + 0.0260 Humidity9am - 0.313 Sunshine
- odds of rain = exp(-1.01 + 0.0260 Humidity9am - 0.313 Sunshine)
- probability of rain = odds / (odds + 1)
Let’s interpret the Sunshine coefficient of -0.313:
## [1] -0.313## [1] 0.7312499- When controlling for humidity, and for every extra hour of sunshine, the LOG(ODDS) of rain…
- decrease by 0.313
- are roughly 31% as big (i.e. decrease by 69%)
- are roughly 73% as big (i.e. decrease by 27%)
- increase by 0.731
- When controlling for humidity, and for every extra hour of sunshine, the ODDS of rain…
- decrease by 0.313
- are roughly 31% as big (i.e. decrease by 69%)
- are roughly 73% as big (i.e. decrease by 27%)
- increase by 0.731
Solution:
- decrease by 0.313
- are roughly 73% as big (i.e. decrease by 27%)
DEFINITION: odds ratio
The coefficients on the odds scale are odds ratios (OR):
\[e^{\beta_1} = \frac{\text{odds of event at x + 1}}{\text{odds of event at x}}\]
EXAMPLE 3: Classifications
log(odds of rain) = -1.01 + 0.0260 Humidity9am - 0.313 Sunshine
Suppose there’s 99% humidity at 9am today and only 2 hours of bright sunshine.
- What’s the probability of rain?
## [1] 0.938- What’s your binary classification: do you predict that it will rain or not rain?
Solution:
- .
## [1] 0.938## [1] 2.554867## [1] 0.7186955- rain
EXAMPLE 4: Classification rules (intuition)
We used a simple classification rule above with a probability threshold of c = 0.5:
- If the probability of rain >= c, then predict rain.
- Otherwise, predict no rain.
Let’s translate this into a classification rule that partitions the data points into rain / no rain predictions based on the predictor values.
What do you think this classification rule / partition will look like?
Solution:
will varyEXAMPLE 5: Building the classification rule
- If …, then predict rain.
- Otherwise, predict no rain.
Work
Identify the pairs of humidity and sunshine values for which the probability of rain is 0.5, hence the log(odds of rain) is 0.
Solution:
Set the log odds to 0:
log(odds of rain) = -1.01 + 0.0260 Humidity9am - 0.313 Sunshine = 0Solve for Humidity9am:
- Move constant and Sunshine term to other side.
0.0260 Humidity9am = 1.01 + 0.3130 Sunshine - Divide both sides by 0.026:
Humidity9am = (1.01 / 0.026) + (0.3130 / 0.026) Sunshine = 38.846 + 12.038 Sunshine
- Move constant and Sunshine term to other side.
EXAMPLE 6: Examine the classification rule
Let’s visualize the partition, hence classification regions defined by our classification rule:
# Example data points
example <- data.frame(Humidity9am = c(90, 90, 60), Sunshine = c(2, 5, 10), RainTomorrow = c(NA, NA, NA))
# Include the line Humidity9am = 38.84615 + 12.03846 Sunshine
rain_plot +
geom_abline(intercept = 38.84615, slope = 12.03846, size = 2) +
geom_point(data = example, color = "red", size = 3)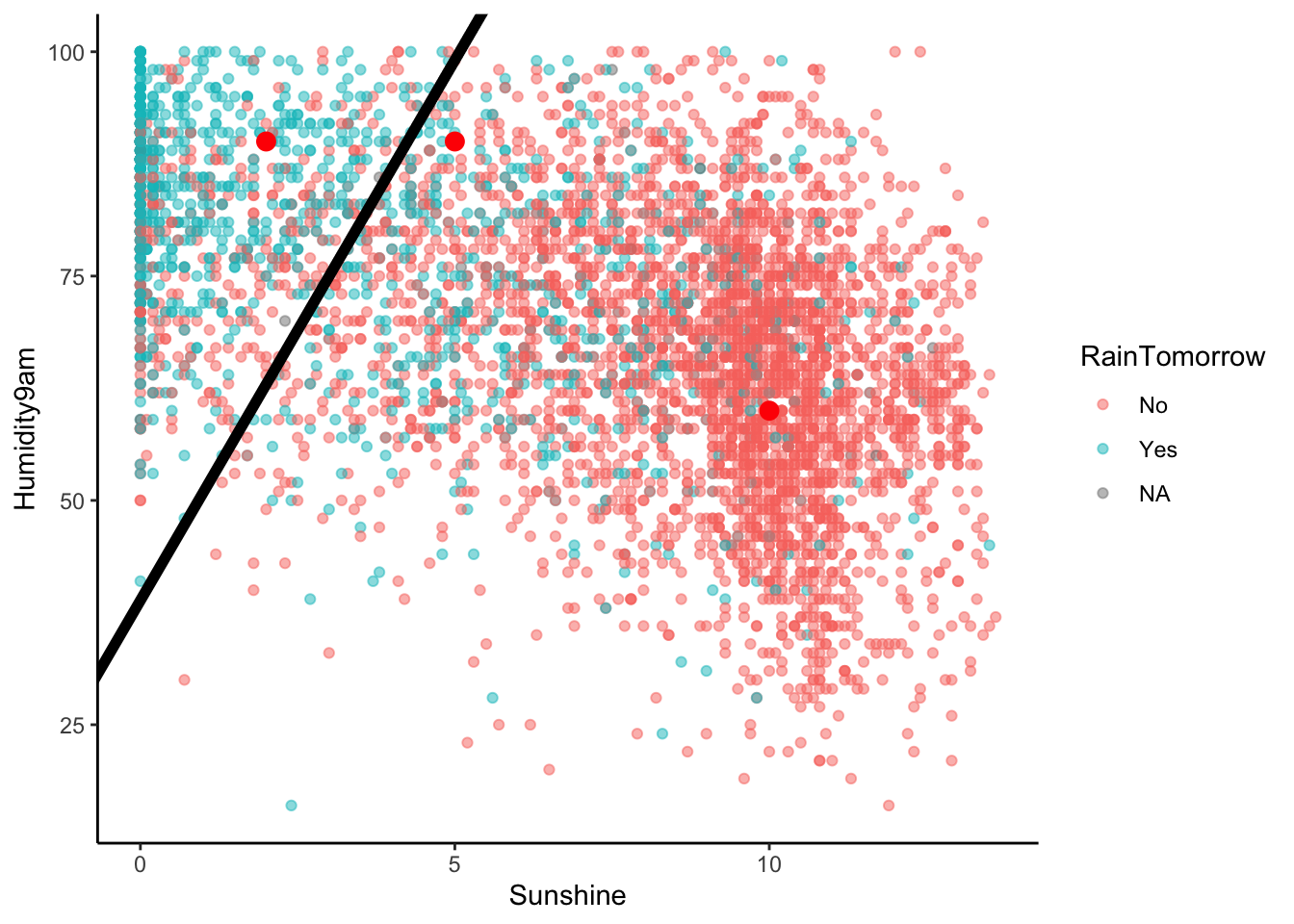
Use our classification rule to predict rain / no rain for the following days:
- Day 1: humidity = 90, sunshine = 2
- Day 2: humidity = 90, sunshine = 5
- Day 3: humidity = 60, sunshine = 10
Solution:
- Day 1: rain
- Day 2: no rain
- Day 3: no rain
EXAMPLE 7: General properties
- Does the logistic regression algorithm have a tuning parameter?
- Estimating the logistic regression model requires the same pre-processing steps as least squares regression.
- Is it necessary to standardize quantitative predictors? If so, does the R function do this for us?
- Is it necessary to create dummy variables for our categorical predictors? If so, does the R function do this for us?
Solution:
- no
- .
- no
- yes and yes
Exercises
Goals
- Implement logistic regression in R.
- Evaluate the accuracy of our logistic regression classifications.
Part 1: Build the model
Let’s continue with our analysis of RainTomorrow vs Humidity9am and Sunshine. You’re given all code here. Be sure to scan and reflect upon what’s happening.
STEP 0: Organize the y categories
We want to model the log(odds of rain), thus the Yes category of RainTomorrow.
But R can’t read minds.
We have to explicitly tell it to treat the No category as the reference level (not the category we want to model).
STEP 1: logistic regression model specification
What’s new here?
STEP 2: variable recipe
There are no required pre-processing steps, but you could add some. Nothing new here!
STEP 3: workflow specification (recipe + model)
Nothing new here!
STEP 4: Estimate the model using the data
Since the logistic regression model has no tuning parameter to tune, we can just fit() the model using our sample data – no need for tune_grid()!
Check out the tidy model summary
Note that these coefficients are the same that we used in the above examples.
## # A tibble: 3 × 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) -1.01 0.258 -3.90 9.61e- 5
## 2 Humidity9am 0.0260 0.00307 8.45 2.91e- 17
## 3 Sunshine -0.313 0.0119 -26.3 2.81e-152# Transform coefficients and confidence intervals to the odds scale
# These are odds ratios (OR)
logistic_model %>%
tidy() %>%
mutate(
OR = exp(estimate),
OR.conf.low = exp(estimate - 1.96*std.error),
OR.conf.high = exp(estimate + 1.96*std.error)
)## # A tibble: 3 × 8
## term estimate std.error statistic p.value OR OR.conf.low
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) -1.01 0.258 -3.90 9.61e- 5 0.365 0.220
## 2 Humidity9am 0.0260 0.00307 8.45 2.91e- 17 1.03 1.02
## 3 Sunshine -0.313 0.0119 -26.3 2.81e-152 0.731 0.714
## # ℹ 1 more variable: OR.conf.high <dbl>Part 2: Apply & evaluate the model
- Predictions & classifications
Consider the weather on 4 days in our data set:
## # A tibble: 4 × 3
## RainTomorrow Humidity9am Sunshine
## <fct> <int> <dbl>
## 1 Yes 75 5.2
## 2 Yes 77 2.1
## 3 Yes 92 3
## 4 No 80 10.1Use the logistic_model to calculate the probability of rain and the rain prediction / classification for these 4 days.
## # A tibble: 4 × 6
## RainTomorrow Humidity9am Sunshine .pred_class .pred_No .pred_Yes
## <fct> <int> <dbl> <fct> <dbl> <dbl>
## 1 Yes 75 5.2 No 0.665 0.335
## 2 Yes 77 2.1 Yes 0.417 0.583
## 3 Yes 92 3 Yes 0.391 0.609
## 4 No 80 10.1 No 0.890 0.110- Convince yourself that you understand what’s being reported in the
.pred_class,.pred_No, and.pred_Yescolumns, as well as the correspondence between these columns (how they’re related to each other). - How many of the 4 classifications were accurate?
Solution:
.pred_class(classification based on probability 0.5 threshold),.pred_No(probability of no rain), and.pred_Yes(probability of rain)- 3
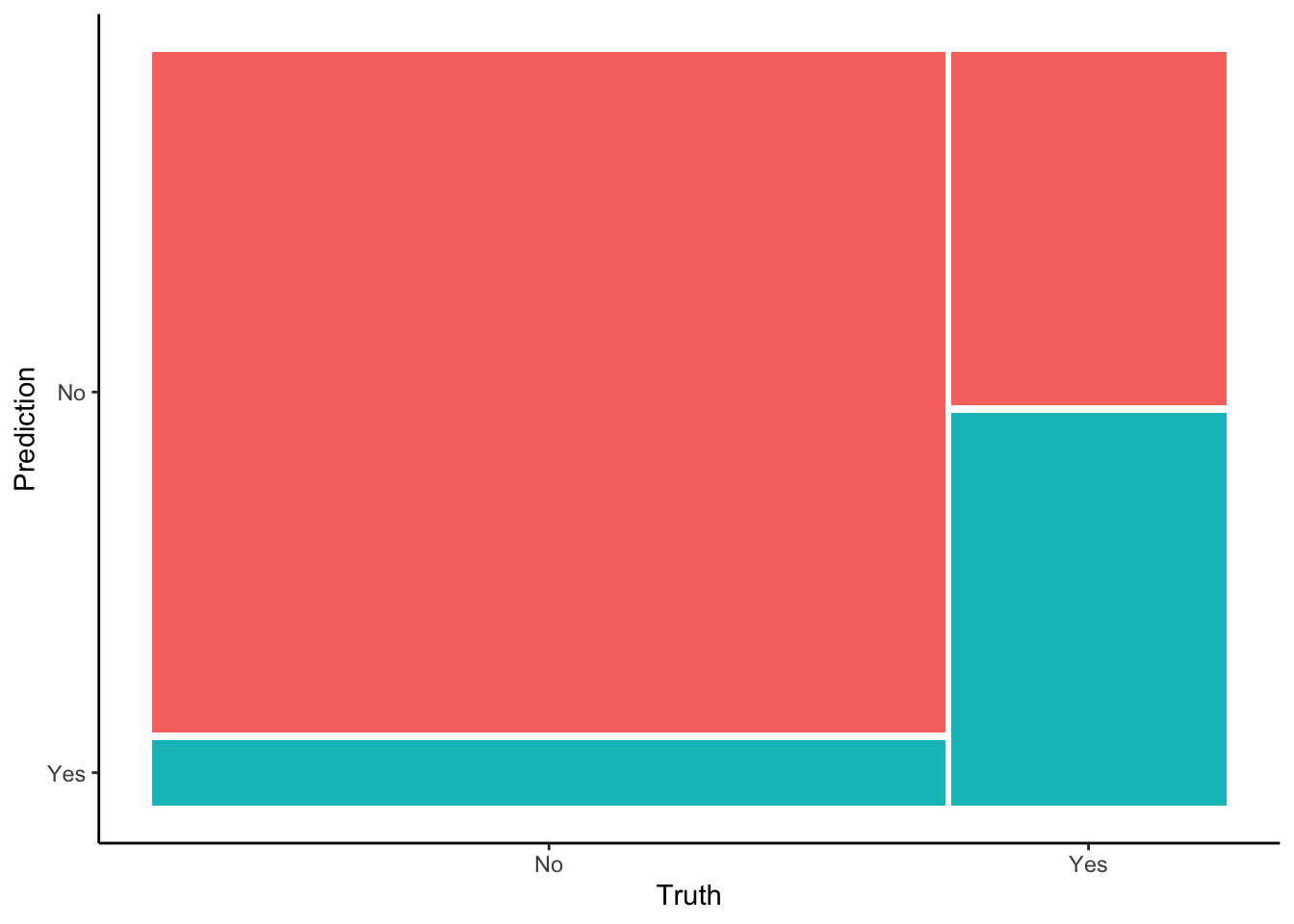
Confusion matrix
Let’s calculate the in_sample_classifications for all days in our sydney sample (“in-sample” because we’re evaluating our model using the same data we used to build it):
in_sample_classifications <- logistic_model %>%
augment(new_data = sydney)
# Check it out
head(in_sample_classifications)## # A tibble: 6 × 6
## RainTomorrow Humidity9am Sunshine .pred_class .pred_No .pred_Yes
## <fct> <int> <dbl> <fct> <dbl> <dbl>
## 1 Yes 92 0 Yes 0.201 0.799
## 2 Yes 83 2.7 Yes 0.425 0.575
## 3 Yes 88 0.1 Yes 0.223 0.777
## 4 Yes 83 0 Yes 0.241 0.759
## 5 Yes 88 0 Yes 0.218 0.782
## 6 Yes 69 8.6 No 0.871 0.129A confusion matrix summarizes the accuracy of the .pred_class model classifications. You’ll answer follow-up questions in the next exercises.
in_sample_confusion <- in_sample_classifications %>%
conf_mat(truth = RainTomorrow, estimate = .pred_class)## Truth
## Prediction No Yes
## No 3119 563
## Yes 301 625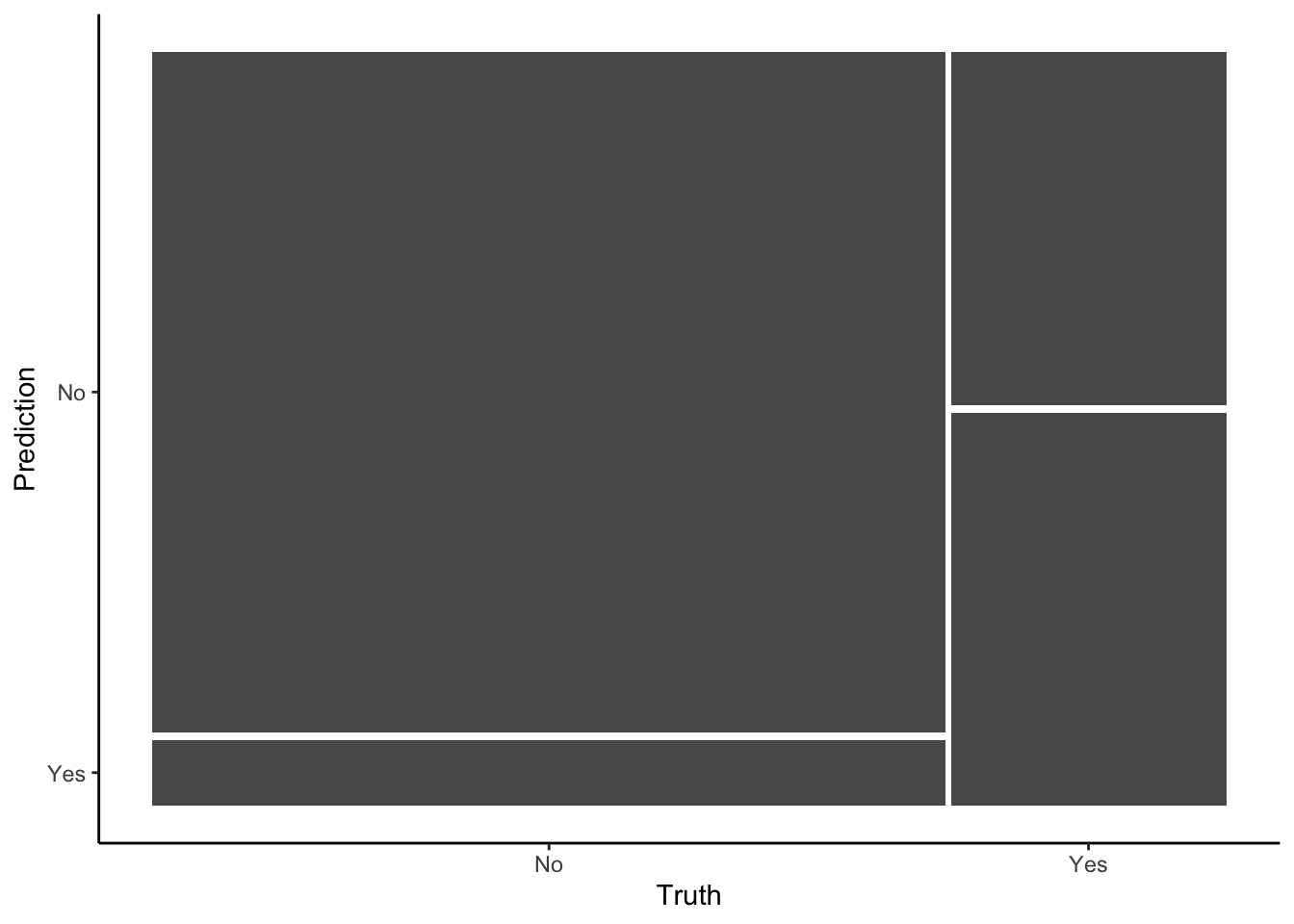
# Check it out in a color plot (which we'll store and use later)
mosaic_plot <- in_sample_confusion %>%
autoplot() +
aes(fill = rep(colnames(in_sample_confusion$table), ncol(in_sample_confusion$table))) +
theme(legend.position = "none")
mosaic_plot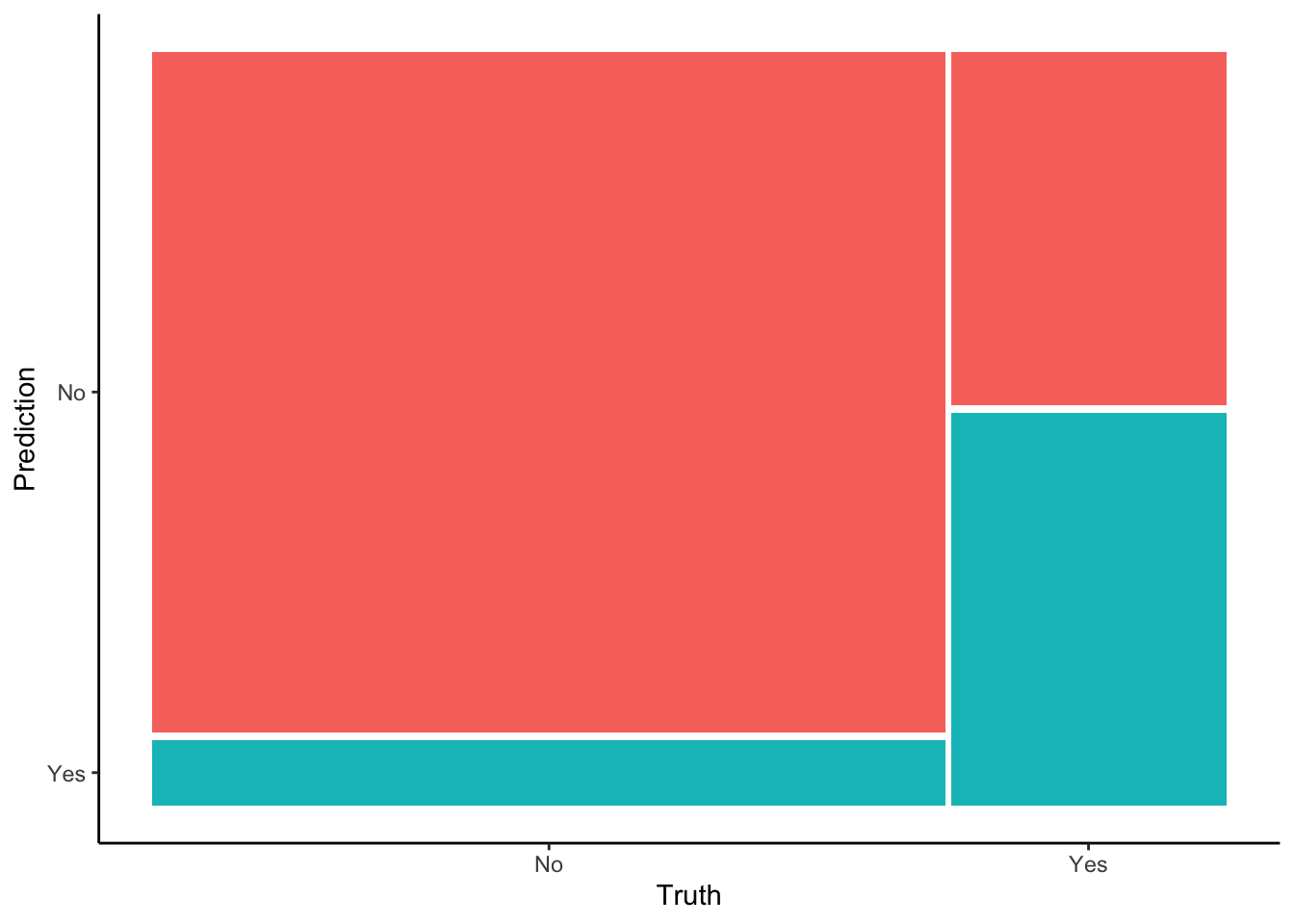
- Overall accuracy
## Truth
## Prediction No Yes
## No 3119 563
## Yes 301 625What do these numbers add up to, both numerically and contextually?
Use this matrix to calculate the overall accuracy of the model classifications. That is, what proportion of the classifications were correct?
Check that your answer to part b matches the
accuracylisted in the confusion matrixsummary():
# event_level indicates that the second RainTomorrow
# category (Yes) is our category of interest
summary(in_sample_confusion, event_level = "second")## # A tibble: 13 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.812
## 2 kap binary 0.472
## 3 sens binary 0.526
## 4 spec binary 0.912
## 5 ppv binary 0.675
## 6 npv binary 0.847
## 7 mcc binary 0.478
## 8 j_index binary 0.438
## 9 bal_accuracy binary 0.719
## 10 detection_prevalence binary 0.201
## 11 precision binary 0.675
## 12 recall binary 0.526
## 13 f_meas binary 0.591Solution:
- sample size (not including data points with NA on variables used in our model)
## [1] 4608## [1] 0.8125- yep
- No information rate
Are our model classifications any better than just randomly guessing rain / no rain?! What if we didn’t even build a model, and just always predicted the most common outcome ofRainTomorrow: that it wouldn’t rain?!
## # A tibble: 3 × 2
## RainTomorrow n
## <fct> <int>
## 1 No 3443
## 2 Yes 1196
## 3 <NA> 14Ignoring the
NAoutcomes, prove that if we just always predicted no rain, we’d be correct 74.2% of the time. This is called the no information rate.Is the overall accuracy of our logistic regression model (81.2%) meaningfully better than this random guessing approach?
Solution:
- We’re only right when it doesn’t rain.
## [1] 0.7421858- this is subjective – depends on context / how we’ll use the predictions / consequences for being wrong.
- Sensitivity
Beyond overall accuracy, we care about the accuracy within each class (rain and no rain). Our model’s true positive rate or sensitivity is the probability that it correctly classifies rain as rain. This is represented by the fraction of rain observations that are red:
# NOTE: We're only plotting RAINY days
in_sample_classifications %>%
filter(RainTomorrow == "Yes") %>%
mutate(correct = (RainTomorrow == .pred_class)) %>%
ggplot(aes(y = Humidity9am, x = Sunshine, color = correct)) +
geom_point(alpha = 0.5) +
geom_abline(intercept = 38.84615, slope = 12.03846, size = 2) +
scale_color_manual(values = c("black", "red"))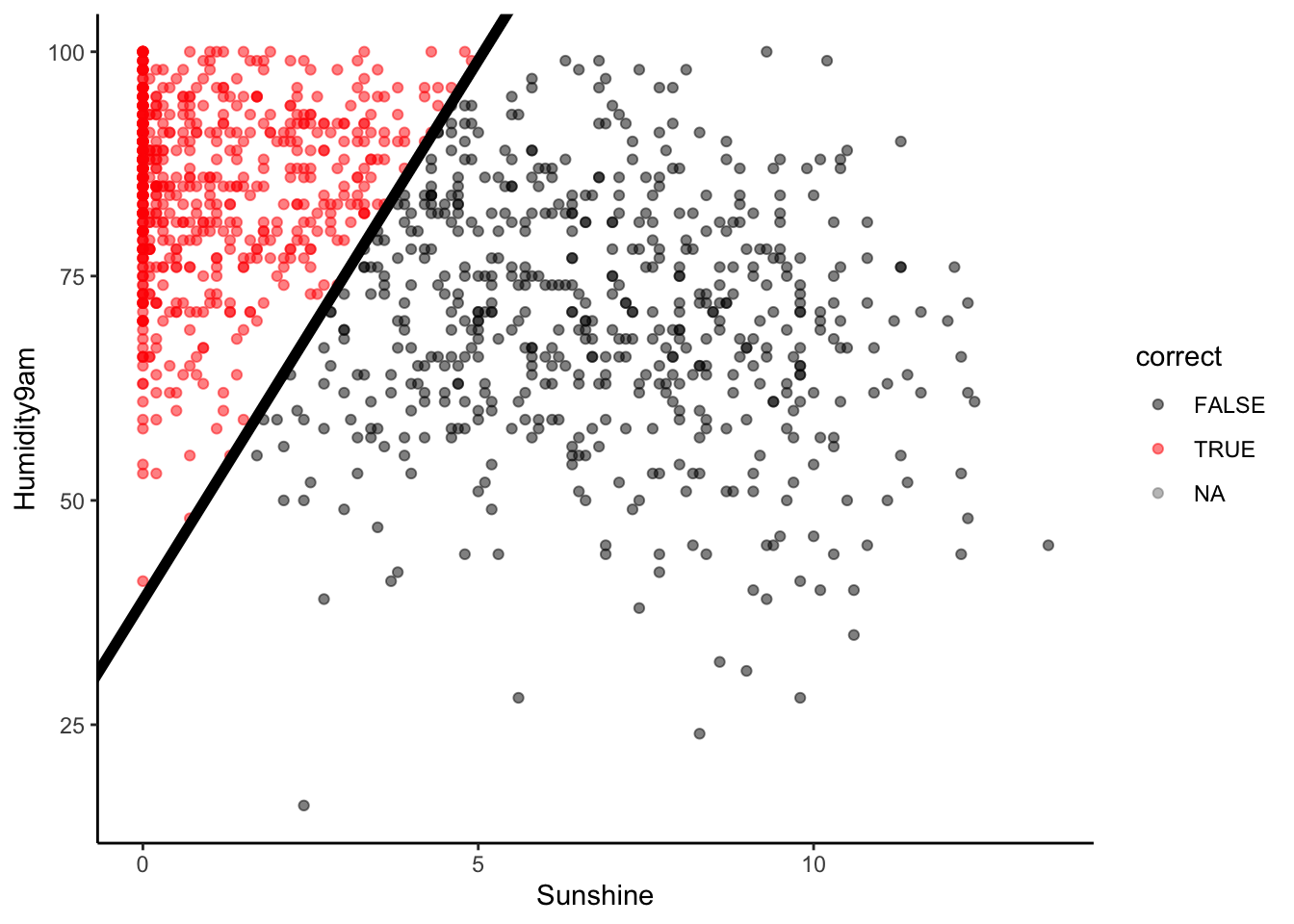
Or, the proportion of the Yes column that falls into the Yes prediction box:
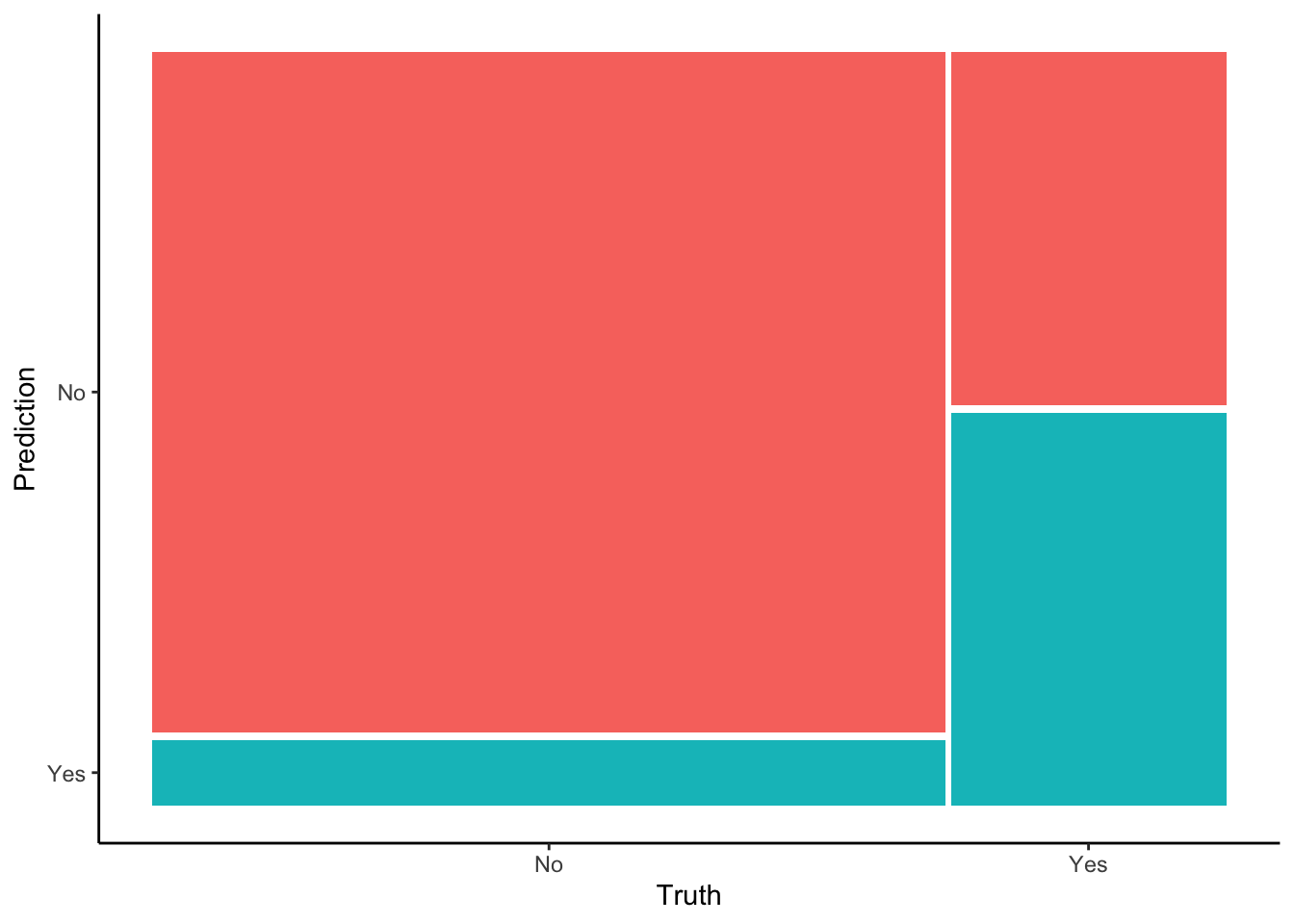
Visually, does it appear that the sensitivity is low, moderate, or high?
Calculate the sensitivity using the confusion matrix.
## Truth
## Prediction No Yes
## No 3119 563
## Yes 301 625- Check that your answer to part b matches the
senslisted in the confusion matrixsummary():
eval=TRUE
## # A tibble: 13 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.812
## 2 kap binary 0.472
## 3 sens binary 0.526
## 4 spec binary 0.912
## 5 ppv binary 0.675
## 6 npv binary 0.847
## 7 mcc binary 0.478
## 8 j_index binary 0.438
## 9 bal_accuracy binary 0.719
## 10 detection_prevalence binary 0.201
## 11 precision binary 0.675
## 12 recall binary 0.526
## 13 f_meas binary 0.591- Interpret the sensitivity and comment on whether this is low, moderate, or high.
Solution:
- moderate (or low)
- .
## [1] 0.5260943- yep
- We correctly anticipate rain 52.6% of the time. Or, on 52.6% of rainy days, we correctly predict rain.
- Specificity
Similarly, we can calculate the model’s true negative rate or specificity, i.e. the probability that it correctly classifies “no rain” as “no rain”. This is represented by the fraction of no rain observations that are red:
# NOTE: We're only plotting NON-RAINY days
in_sample_classifications %>%
filter(RainTomorrow == "No") %>%
mutate(correct = (RainTomorrow == .pred_class)) %>%
ggplot(aes(y = Humidity9am, x = Sunshine, color = correct)) +
geom_point(alpha = 0.5) +
geom_abline(intercept = 38.84615, slope = 12.03846, size = 2) +
scale_color_manual(values = c("black", "red"))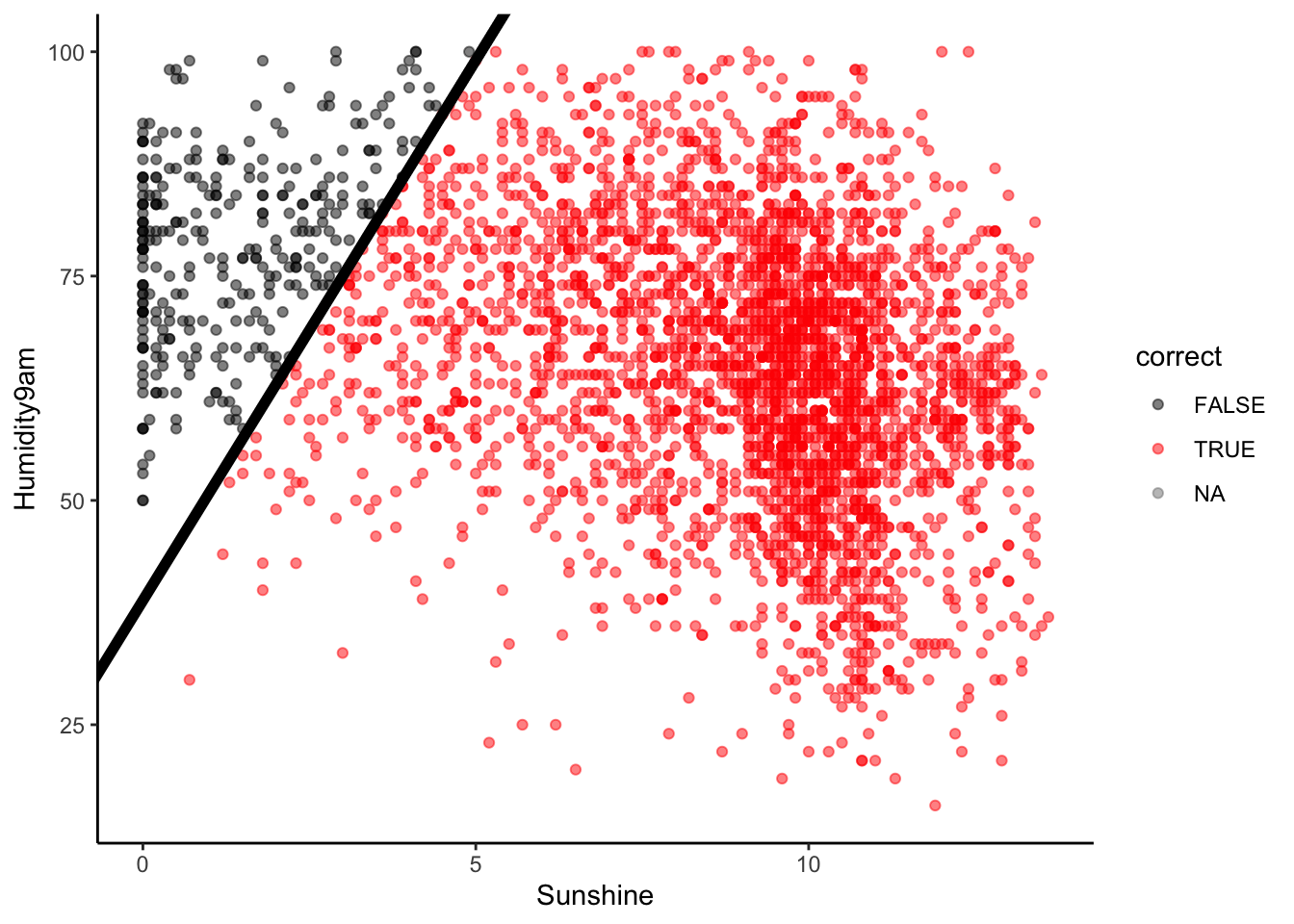
Or, the proportion of the No column that falls into the No prediction box:
Visually, does it appear that the specificity is low, moderate, or high?
Calculate specificity using the confusion matrix.
## Truth
## Prediction No Yes
## No 3119 563
## Yes 301 625- Check that your answer to part b matches the
speclisted in the confusion matrixsummary():
## # A tibble: 13 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.812
## 2 kap binary 0.472
## 3 sens binary 0.526
## 4 spec binary 0.912
## 5 ppv binary 0.675
## 6 npv binary 0.847
## 7 mcc binary 0.478
## 8 j_index binary 0.438
## 9 bal_accuracy binary 0.719
## 10 detection_prevalence binary 0.201
## 11 precision binary 0.675
## 12 recall binary 0.526
## 13 f_meas binary 0.591- Interpret the specificity and comment on whether this is low, moderate, or high.
Solution:
- high
- .
## [1] 0.9119883- yep
## # A tibble: 13 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.812
## 2 kap binary 0.472
## 3 sens binary 0.526
## 4 spec binary 0.912
## 5 ppv binary 0.675
## 6 npv binary 0.847
## 7 mcc binary 0.478
## 8 j_index binary 0.438
## 9 bal_accuracy binary 0.719
## 10 detection_prevalence binary 0.201
## 11 precision binary 0.675
## 12 recall binary 0.526
## 13 f_meas binary 0.591- On 91.2% of non-rainy days, we correctly predict no rain.
- In-sample vs CV Accuracy
The above in-sample metrics of overall accuracy (0.812), sensitivity (0.526), and specificity (0.912) helped us understand how well our model classifies rain / no rain for the same data points we used to build the model. Let’s calculate the cross-validated metrics to better understand how well our model might classify days in the future:
# NOTE: This is very similar to the code for CV with least squares!
# EXCEPT: We need the "control" argument to again specify our interest in the "Yes" category
set.seed(253)
logistic_model_cv <- logistic_spec %>%
fit_resamples(
RainTomorrow ~ Humidity9am + Sunshine,
resamples = vfold_cv(sydney, v = 10),
control = control_resamples(save_pred = TRUE, event_level = 'second'),
metrics = metric_set(accuracy, sensitivity, specificity)
)
# Check out the resulting CV metrics
logistic_model_cv %>%
collect_metrics()## # A tibble: 3 × 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 accuracy binary 0.812 10 0.00651 Preprocessor1_Model1
## 2 sensitivity binary 0.527 10 0.0149 Preprocessor1_Model1
## 3 specificity binary 0.911 10 0.00677 Preprocessor1_Model1How similar are the in-sample and CV evaluation metrics? Based on these, do you think our model is overfit?
Solution:
They’re similar, thus our model doesn’t seem overfit.- Specificity vs Sensitivity
- Our model does better at correctly predicting non-rainy days than rainy days (specificity > sensitivity). Why do you think this is the case?
- In the context of predicting rain, what would you prefer: high sensitivity or high specificity?
- Changing up the probability threshold we use in classifying days as rain / no rain gives us some control over sensitivity and specificity. Consider lowering the threshold from 0.5 to 0.05. Thus if there’s even a 5% chance of rain, we’ll predict rain! What’s your intuition:
- sensitivity will decrease and specificity will increase
- sensitivity will increase and specificity will decrease
- both sensitivity and specificity will increase
Solution:
- because non-rainy days are much more common
- will vary. would you rather risk getting wet instead of carrying your umbrella, or carry your umbrella when it doesn’t rain?
- will vary
- Change up the threshold
Let’s try lowering the threshold to 0.05!
# Calculate .pred_class using a 0.05 threshold
# (this overwrites the default .pred_class which uses 0.5)
new_classifications <- logistic_model %>%
augment(new_data = sydney) %>%
mutate(.pred_class = ifelse(.pred_Yes >= 0.05, "Yes", "No")) %>%
mutate(.pred_class = as.factor(.pred_class))# Obtain a new confusion matrix
new_confusion <- new_classifications %>%
conf_mat(truth = RainTomorrow, estimate = .pred_class)
new_confusion## Truth
## Prediction No Yes
## No 622 23
## Yes 2798 1165## # A tibble: 13 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.388
## 2 kap binary 0.0922
## 3 sens binary 0.981
## 4 spec binary 0.182
## 5 ppv binary 0.294
## 6 npv binary 0.964
## 7 mcc binary 0.205
## 8 j_index binary 0.163
## 9 bal_accuracy binary 0.581
## 10 detection_prevalence binary 0.860
## 11 precision binary 0.294
## 12 recall binary 0.981
## 13 f_meas binary 0.452How does the new sensitivity compare to that using the 0.5 threshold (0.526)?
How does the new specificity compare to that using the 0.5 threshold (0.912)?
Was your intuition right? When we decrease the probability threshold…
- sensitivity decreases and specificity increases
- sensitivity increases and specificity decreases
- both sensitivity and specificity increase
- WE get to pick an appropriate threshold for our analysis. Change up 0.05 in the code below to identify a threshold you like.
# Calculate .pred_class using a 0.05 threshold
# (this overwrites the defaulty .pred_class which uses 0.5)
new_classifications <- logistic_model %>%
augment(new_data = sydney) %>%
mutate(.pred_class = ifelse(.pred_Yes >= 0.05, "Yes", "No")) %>%
mutate(.pred_class = as.factor(.pred_class))
# Obtain a new confusion matrix
new_confusion <- new_classifications %>%
conf_mat(truth = RainTomorrow, estimate = .pred_class)
new_confusion
# Obtain new summaries
summary(new_confusion, event_level = "second")Solution:
- much higher (0.981)
- much lower (0.182)
- sensitivity increases and specificity decreases
- will vary
- OPTIONAL challenge
In Example 5, we built the following classification rule based on a 0.5 probability threshold:
- If
Humidity9am > 38.84615 + 12.03846 Sunshine, then predict rain. - Otherwise, predict no rain.
And we plotted this rule:
ggplot(sydney, aes(y = Humidity9am, x = Sunshine, color = RainTomorrow)) +
geom_point(alpha = 0.5) +
geom_abline(intercept = 38.84615, slope = 12.03846, size = 2) +
geom_point(data = example, color = "black", size = 3)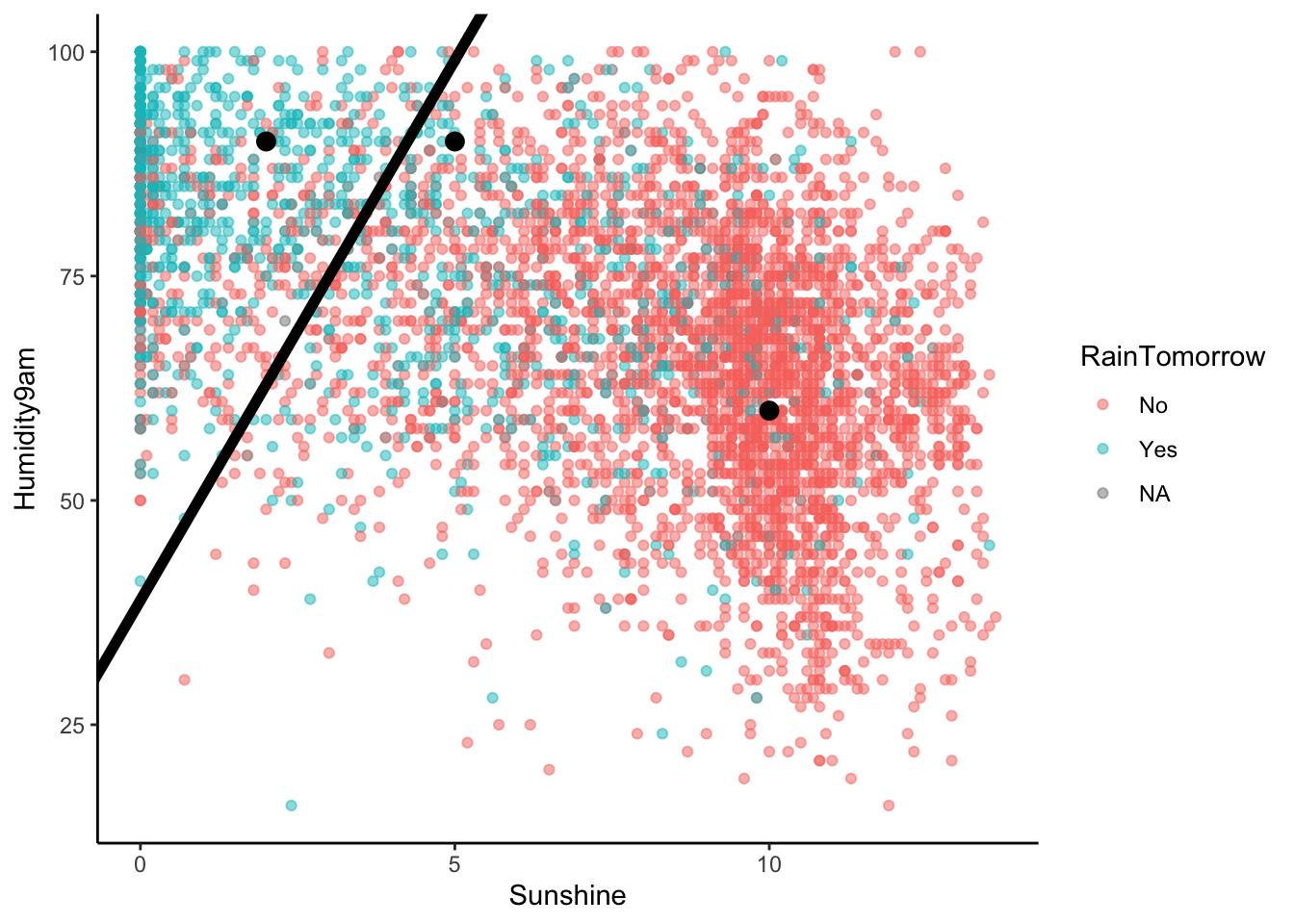
Challenge: Modify this rule and the plot using a 0.05 probability threshold.
Solution:
- If
Humidity9am > -74.385 + 12.03846 Sunshine, then predict rain. - Otherwise, predict no rain.
ggplot(sydney, aes(y = Humidity9am, x = Sunshine, color = RainTomorrow)) +
geom_point(alpha = 0.5) +
geom_abline(intercept = -74.385, slope = 12.03846, size = 2) +
geom_point(data = example, color = "black", size = 3)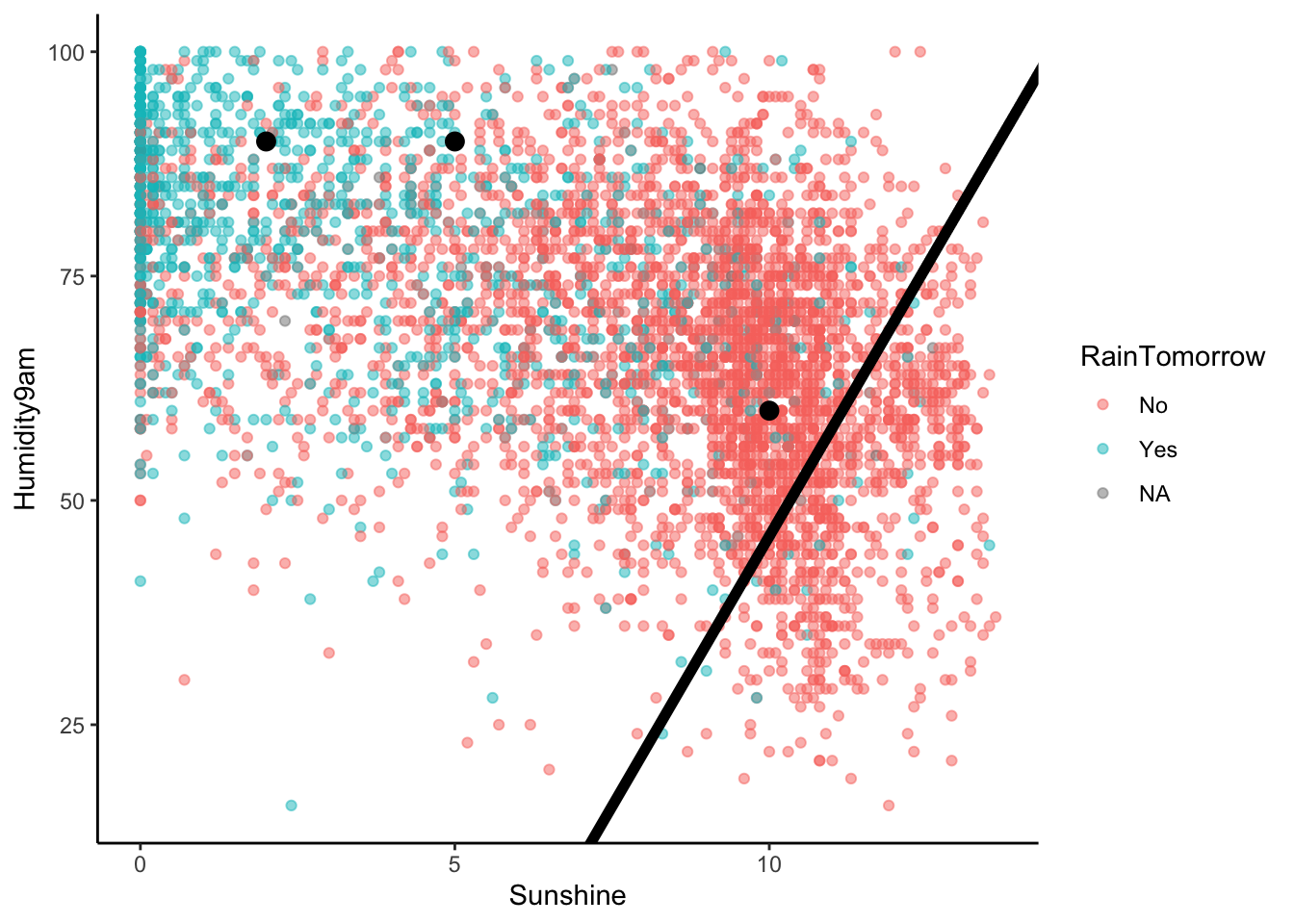
Work
## [1] -2.944439Set log(odds) to -2.944:
log(odds of rain) = -1.01 + 0.0260 Humidity9am - 0.313 Sunshine = -2.944
0.0260 Humidity9am = -2.944 + 1.01 + 0.3130 Sunshine = -1.934 + 0.3130 Sunshine
Humidity9am = (-1.934/0.0260) + (0.3130/0.0260) Sunshine = -74.385 + 12.038 Sunshine
- OPTIONAL math
For a general logistic regression model
\[log(\text{odds}) = \beta_0 + \beta_1 x\]
\(\beta_1\) is the change in log(odds) when we increase \(x\) by 1:
\[\beta_1 = log(\text{odds at x + 1}) - log(\text{odds at x})\]
Prove \(e^{\beta_1}\) is the multiplicative change in odds when we increase \(x\) by 1.
Solution:
\[\begin{split} \beta_1 & = log(\text{odds at x + 1}) - log(\text{odds at x}) \\ & = log\left(\frac{\text{odds at x + 1}}{\text{odds at x}} \right) \\ e^{\beta_1} & = e^{log\left(\frac{\text{odds at x + 1}}{\text{odds at x}} \right)}\\ & = \frac{\text{odds at x + 1}}{\text{odds at x}} \\ \end{split}\]Deeper learning (OPTIONAL)
Recall that in least squares regression, we use residuals to both estimate model coefficients (those that minimize the residual sum of squares) and measure model strength (\(R^2\) is calculated from the variance of the residuals). BUT the concept of a “residual” is different in logistic regression. Mainly, we observe binary y outcomes but our predictions are on the probability scale. In this case, logistic regression requires different strategies for estimating and evaluating models.
Calculating coefficient estimates
A common strategy is to use iterative processes to identify coefficient estimates \(\hat{\beta}\) that maximize the likelihood function \[L(\hat{\beta}) = \prod_{i=1}^{n} p_i^{y_i}(1-p_i)^{1-y_i} \;\; \text{ where } \;\; log\left(\frac{p_i}{1-p_i}\right) = \hat{\beta}_0 + \hat{\beta}_1 x\]Measuring model quality
Akaike’s Information Criterion (AIC) is a common metric with which to compare models. The smaller the AIC the better! Specifically: \[\text{AIC} = \text{-(likelihood of our model)} + 2(p + 1)\] where \(p\) is the number of non-intercept coefficients.
Notes: R code
Suppose we want to build a model of categorical response variable y using predictors x1 and x2 in our sample_data.
# Load packages
library(tidymodels)
# Resolves package conflicts by preferring tidymodels functions
tidymodels_prefer()Organize the y categories
Unless told otherwise, our R functions will model the log(odds) of whatever y category is last alphabetically.
To be safe, we should always set the reference level of y to the outcome we are NOT interested in (eg: “No” if modeling RainTomorrow).
Build the model
# STEP 1: logistic regression model specification
logistic_spec <- logistic_reg() %>%
set_mode("classification") %>%
set_engine("glm")# STEP 2: variable recipe
# There are no REQUIRED pre-processing steps, but you CAN add some
variable_recipe <- recipe(y ~ x1 + x2, data = sample_data)# STEP 3: workflow specification (recipe + model)
logistic_workflow <- workflow() %>%
add_recipe(variable_recipe) %>%
add_model(logistic_spec) # STEP 4: Estimate the model using the data
logistic_model <- logistic_workflow %>%
fit(data = sample_data)Examining model coefficients
# Get a summary table
logistic_model %>%
tidy()
# Transform coefficients and confidence intervals to the odds scale
# These are odds ratios (OR)
logistic_model %>%
tidy() %>%
mutate(
OR = exp(estimate),
OR.conf.low = exp(estimate - 1.96*std.error),
OR.conf.high = exp(estimate + 1.96*std.error)
)Calculate predictions and classifications
# augment gives both probability calculations and classifications
# Plug in a data.frame object with observations on each predictor in the model
logistic_model %>%
augment(new_data = ___)
# We can also use predict!
# Make soft (probability) predictions
logistic_model %>%
predict(new_data = ___, type = "prob")
# Make hard (class) predictions (using a default 0.5 probability threshold)
logistic_model %>%
predict(new_data = ___, type = "class")In-sample evaluation metrics
# Calculate in-sample classifications
in_sample_classifications <- logistic_model %>%
augment(new_data = sample_data)
# Confusion matrix
in_sample_confusion <- in_sample_classifications %>%
conf_mat(truth = y, estimate = .pred_class)
# Summaries
# event_level = "second" indicates that the second category
# is the category of interest
summary(in_sample_confusion, event_level = "second")
# Mosaic plots
in_sample_confusion %>%
autoplot()
# Mosaic plot with color
in_sample_confusion %>%
autoplot() +
aes(fill = rep(colnames(in_sample_confusion$table), ncol(in_sample_confusion$table))) +
theme(legend.position = "none")Cross-validated evaluation metrics
set.seed(___)
logistic_model_cv <- logistic_spec %>%
fit_resamples(
y ~ x1 + x2,
resamples = vfold_cv(sample_data, v = ___),
control = control_resamples(save_pred = TRUE, event_level = 'second'),
metrics = metric_set(accuracy, sensitivity, specificity)
)
# Check out the resulting CV metrics
logistic_model_cv %>%
collect_metrics()